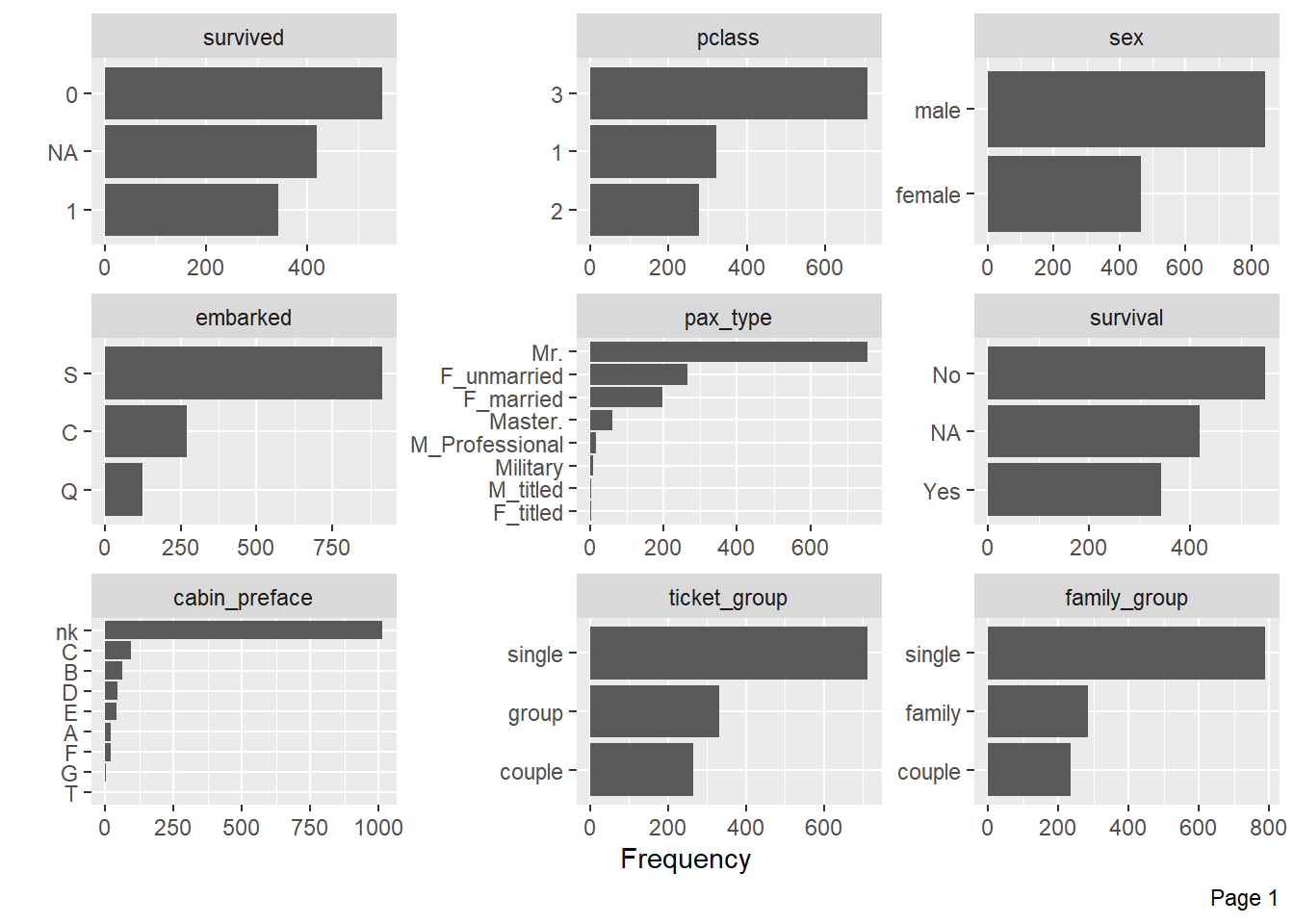
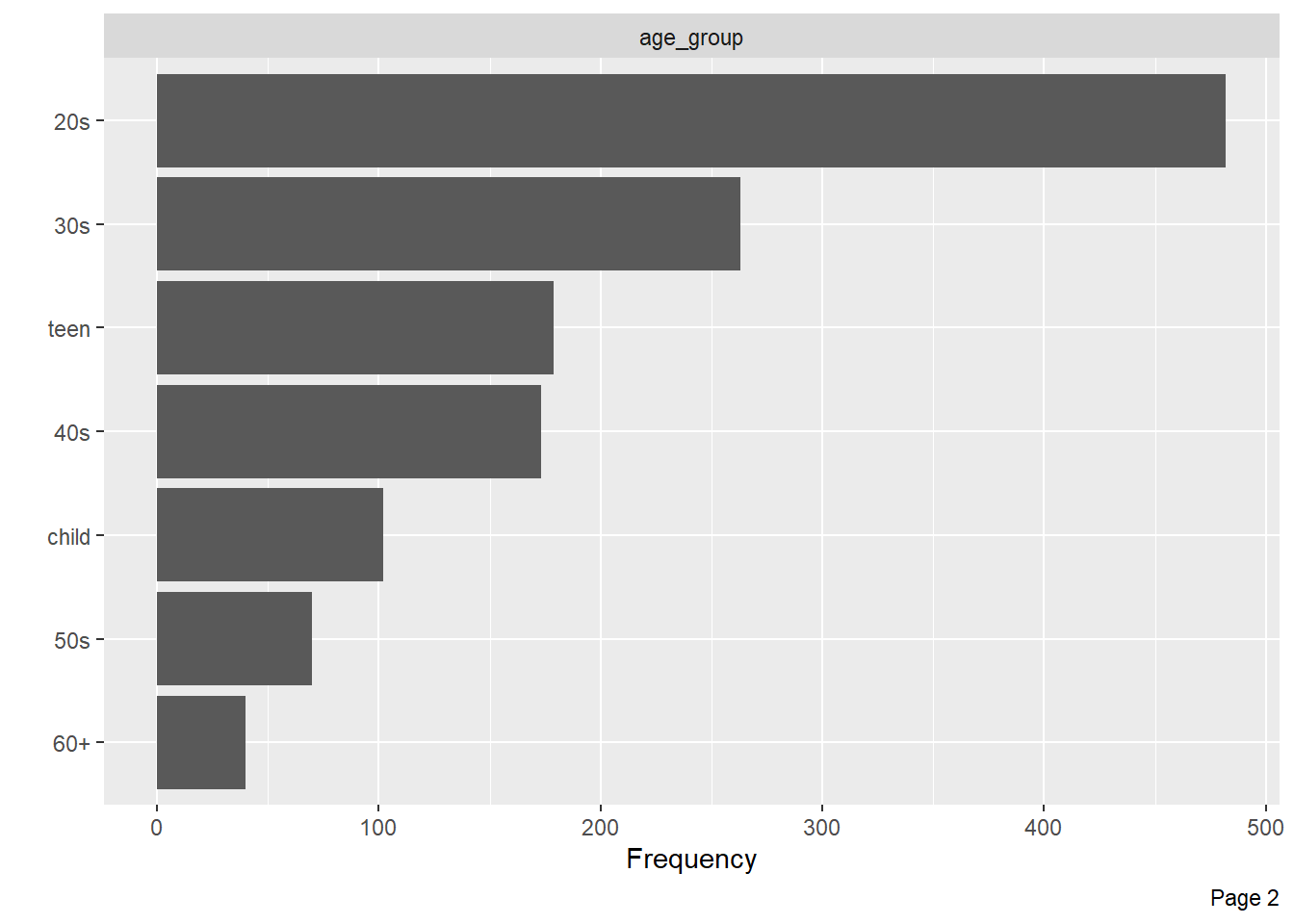
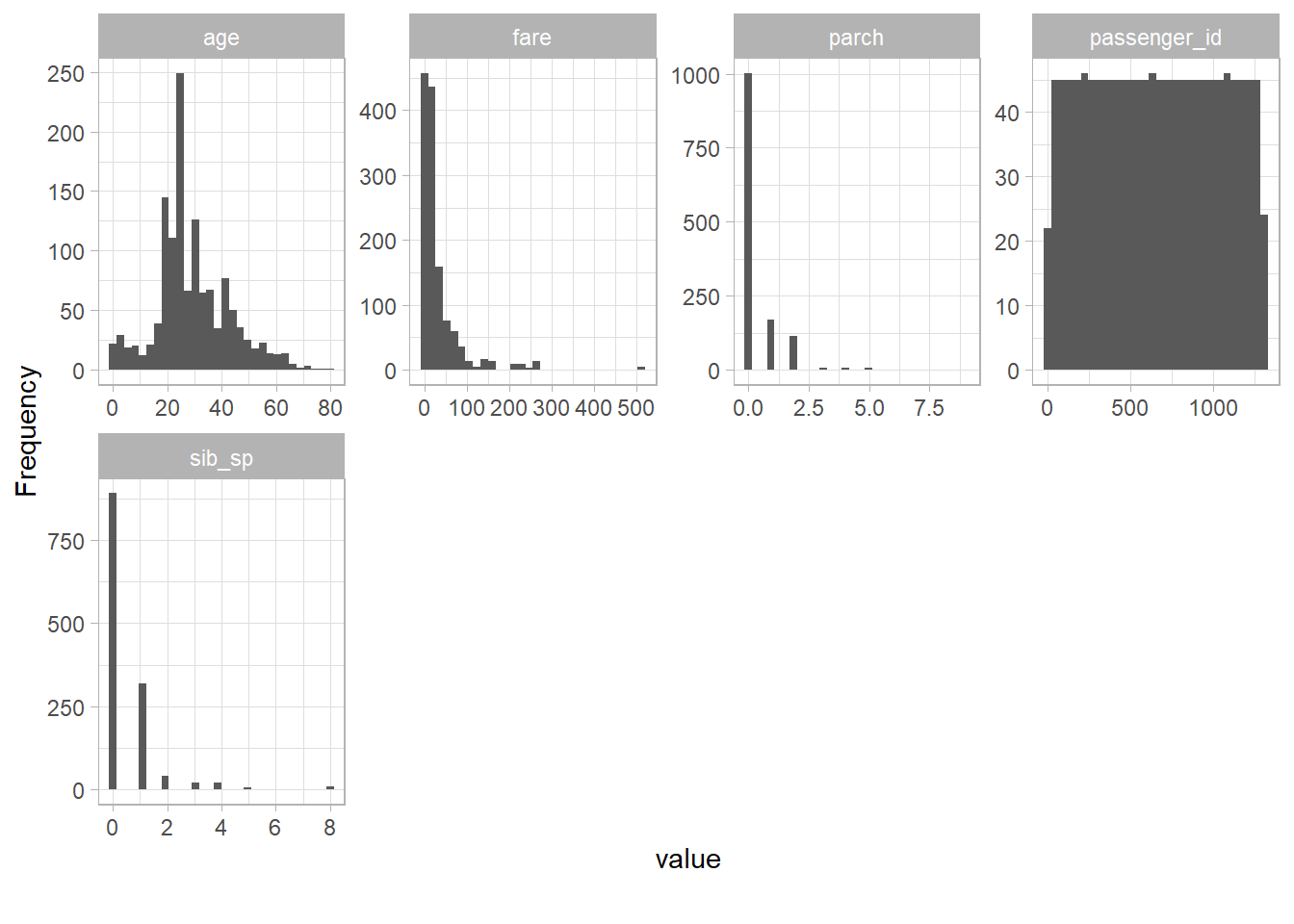
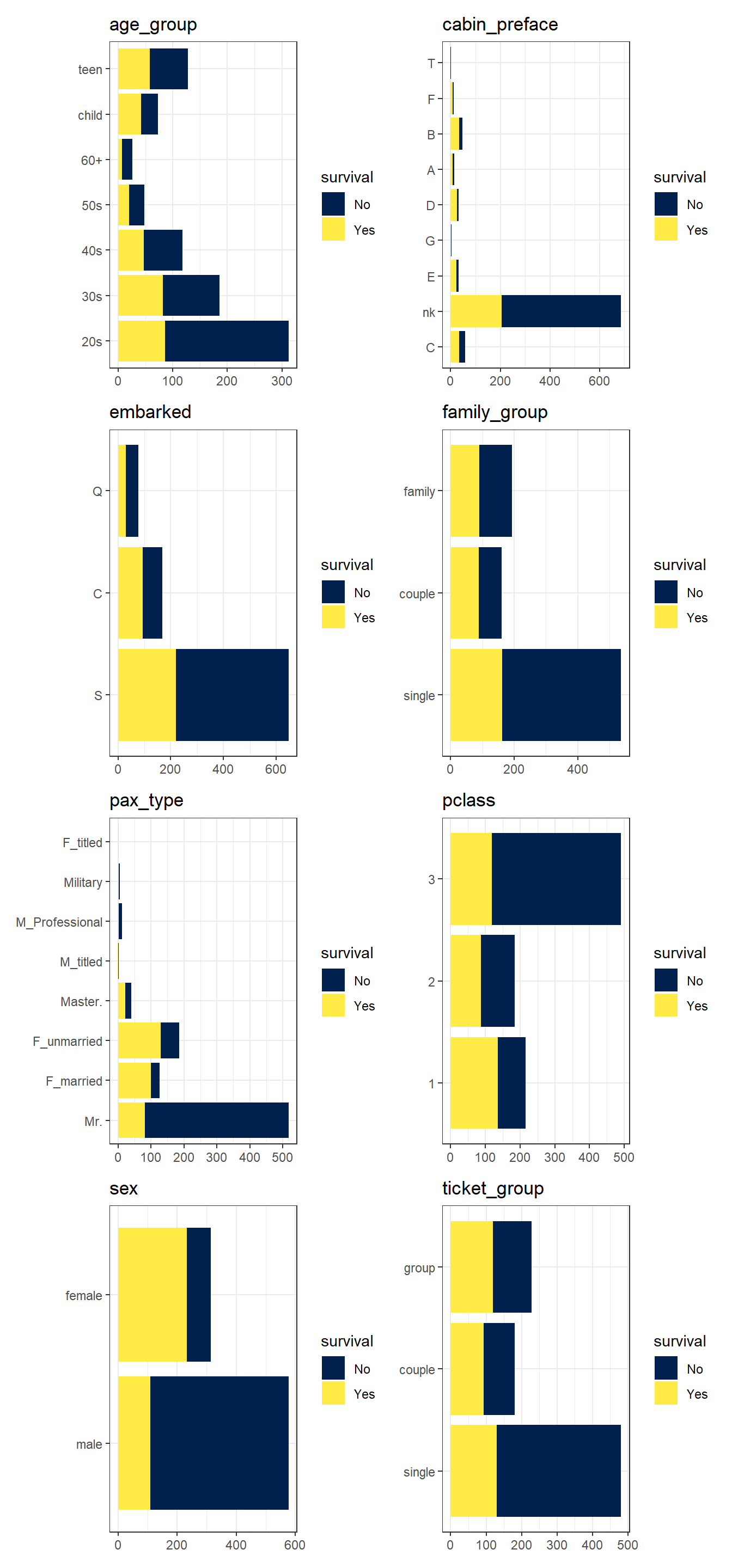
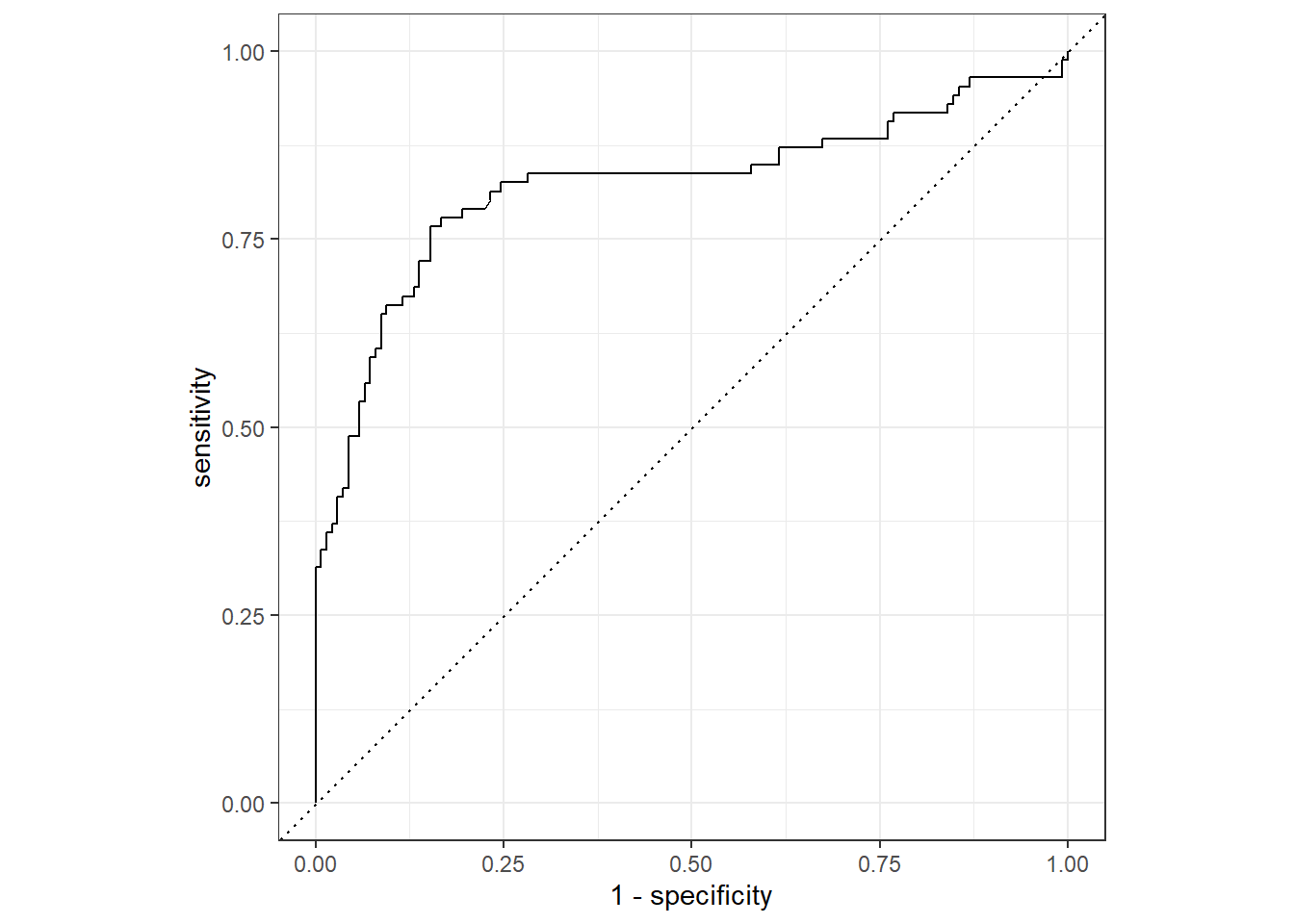
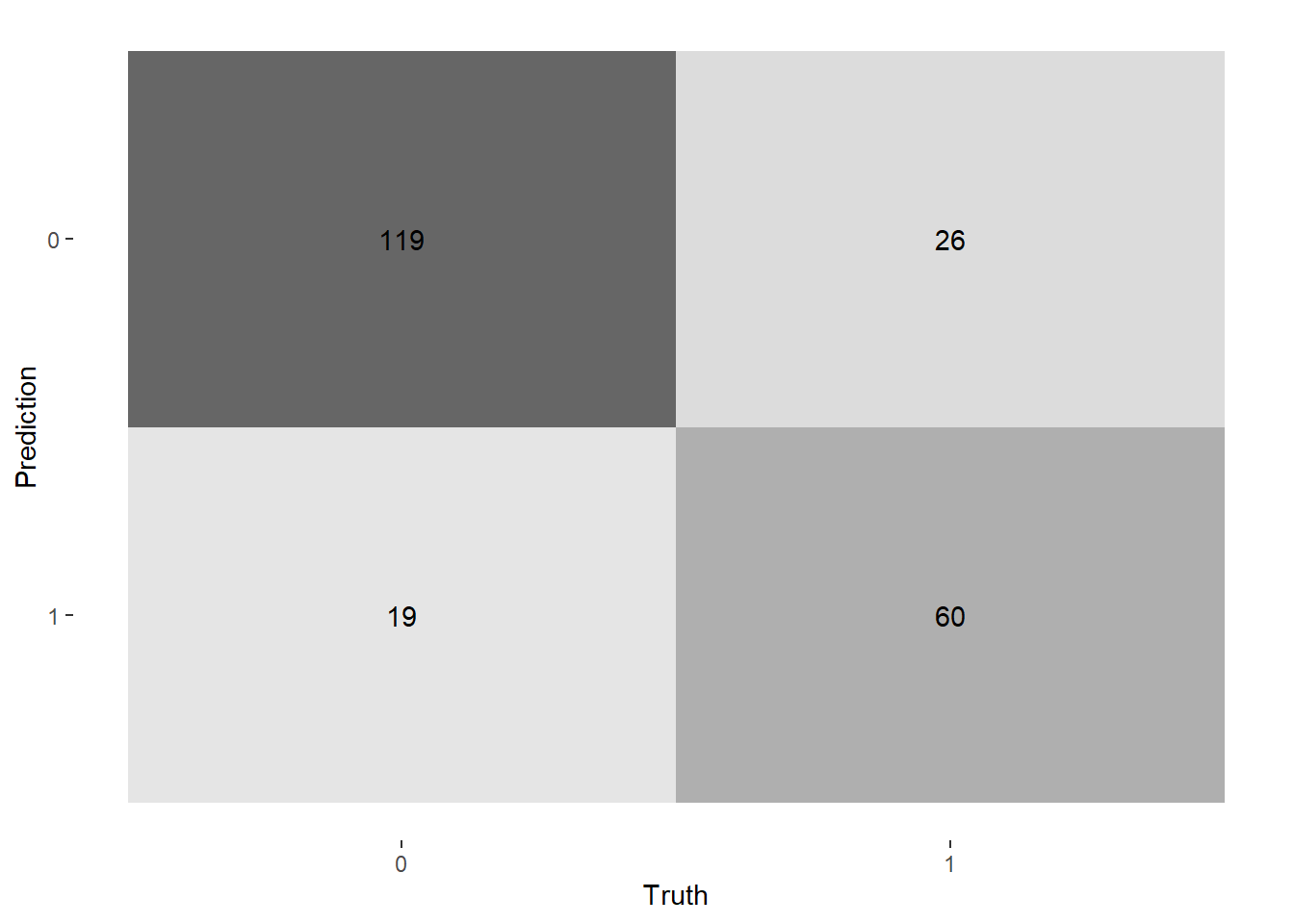
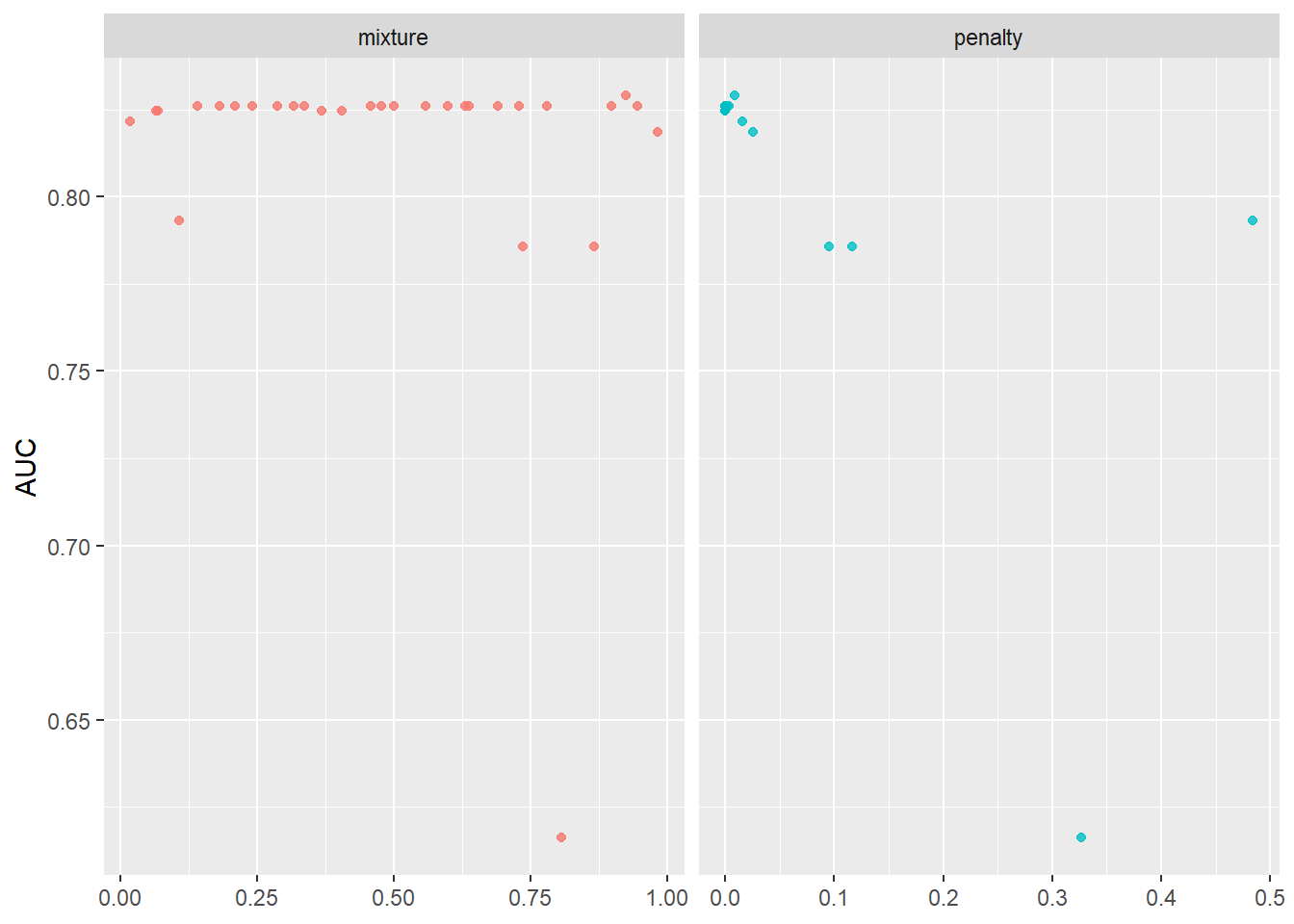
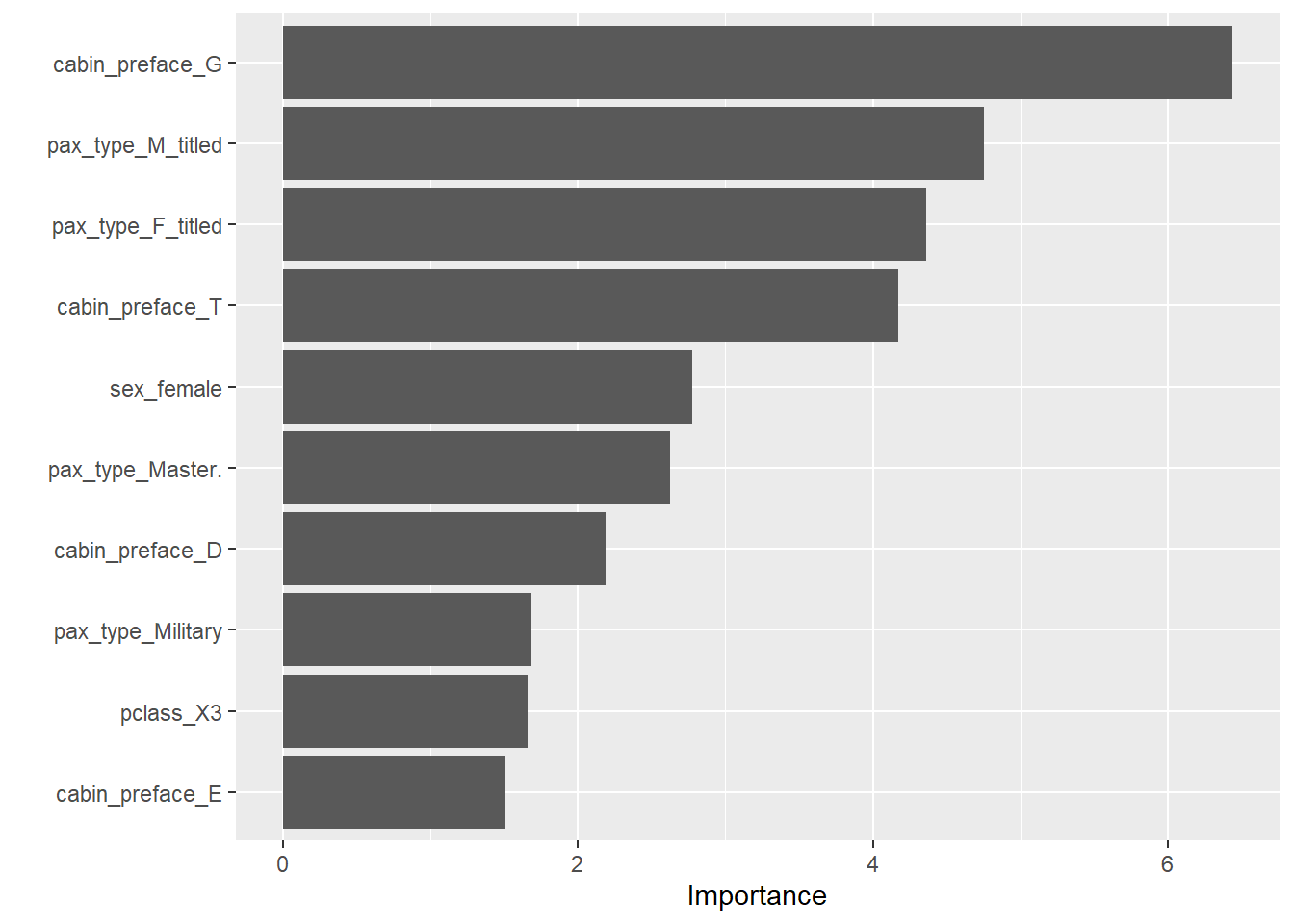
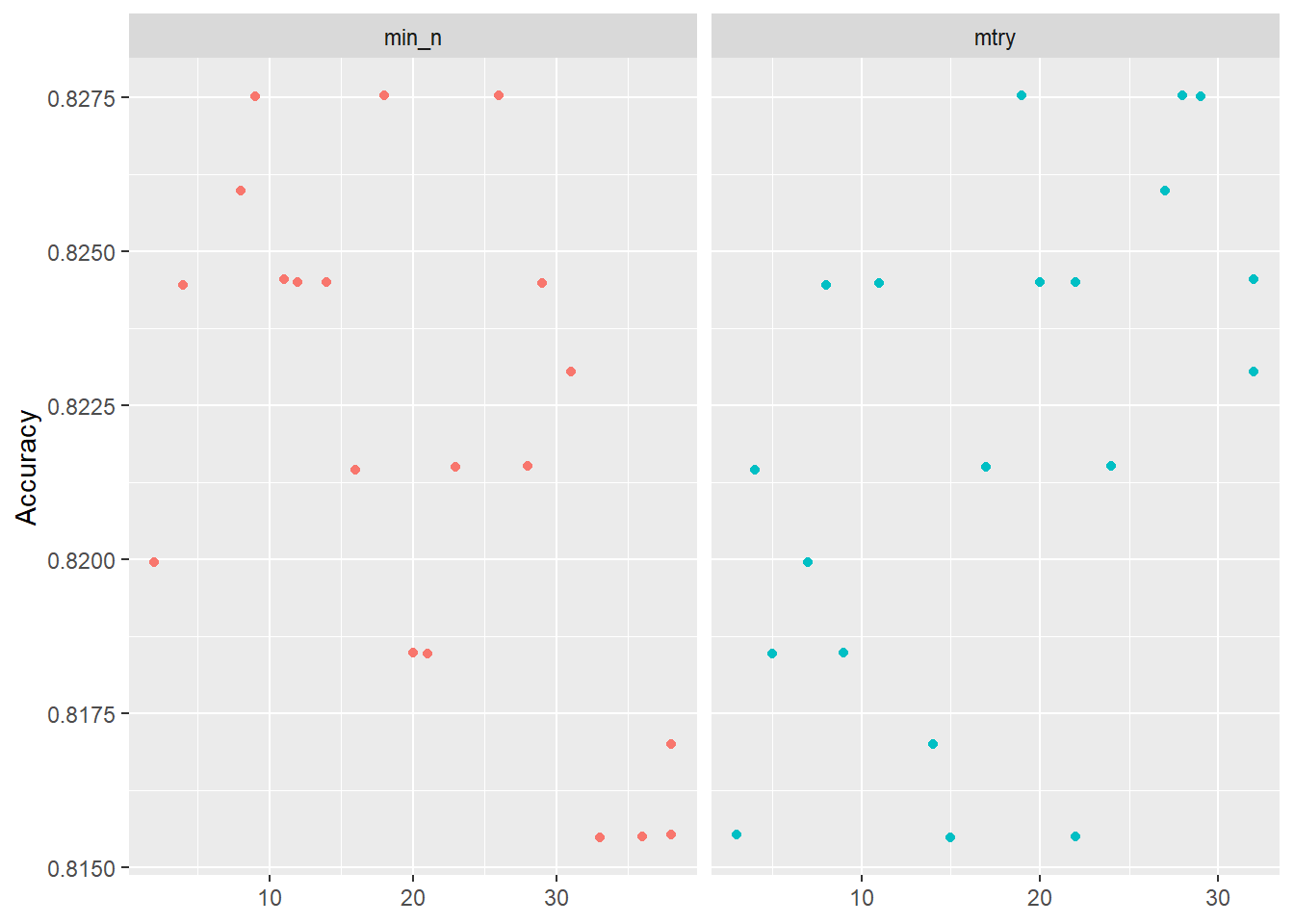
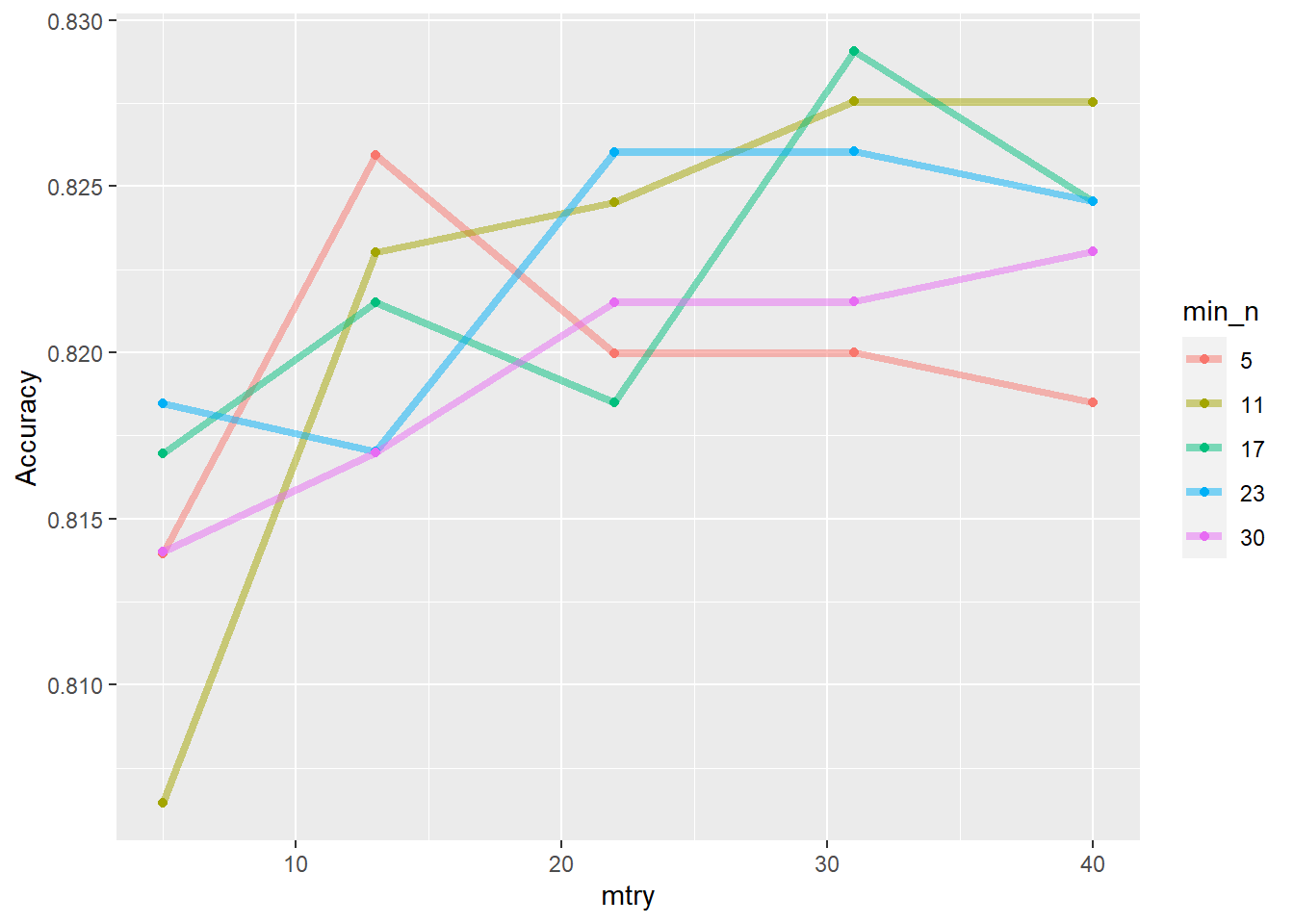
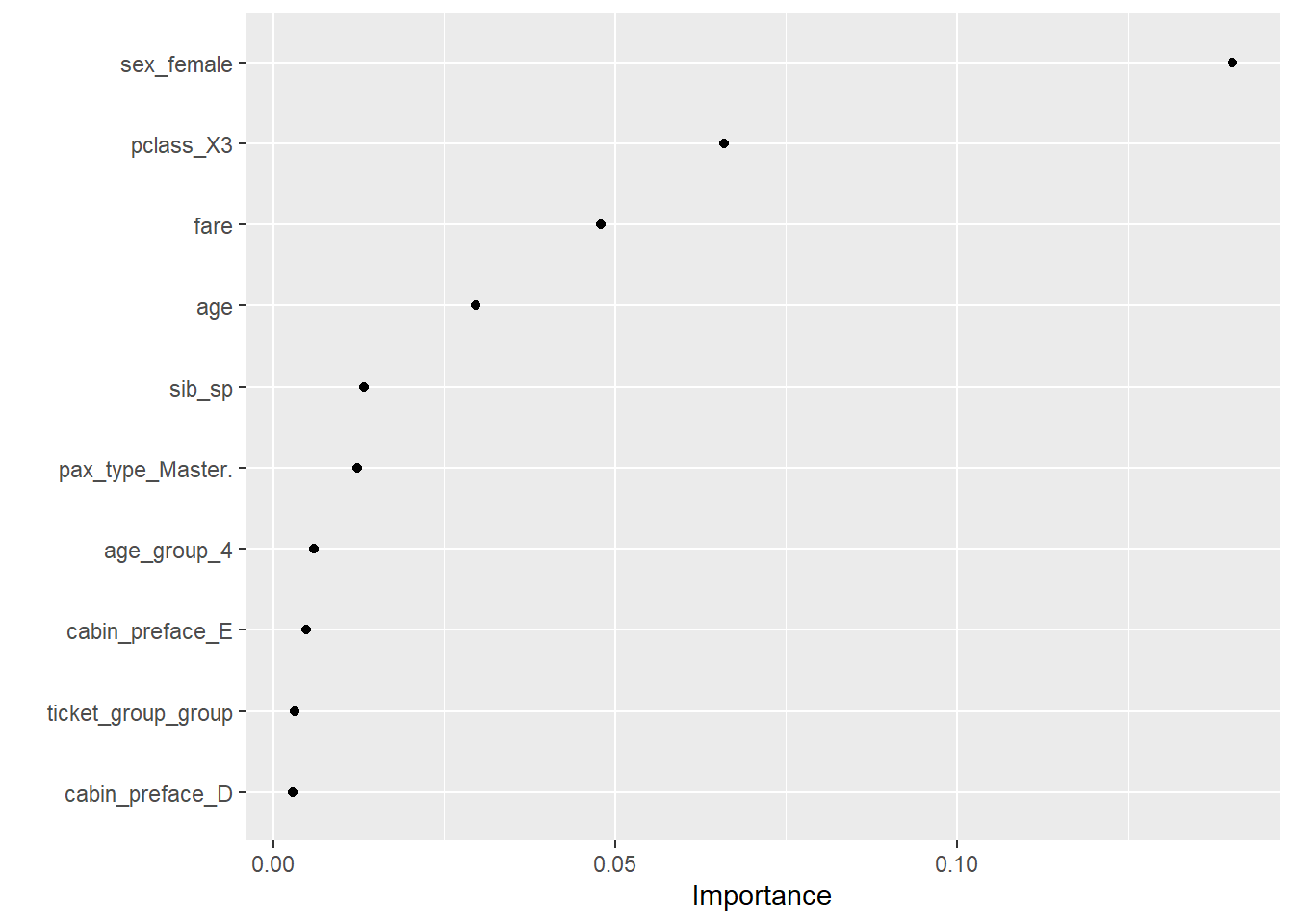
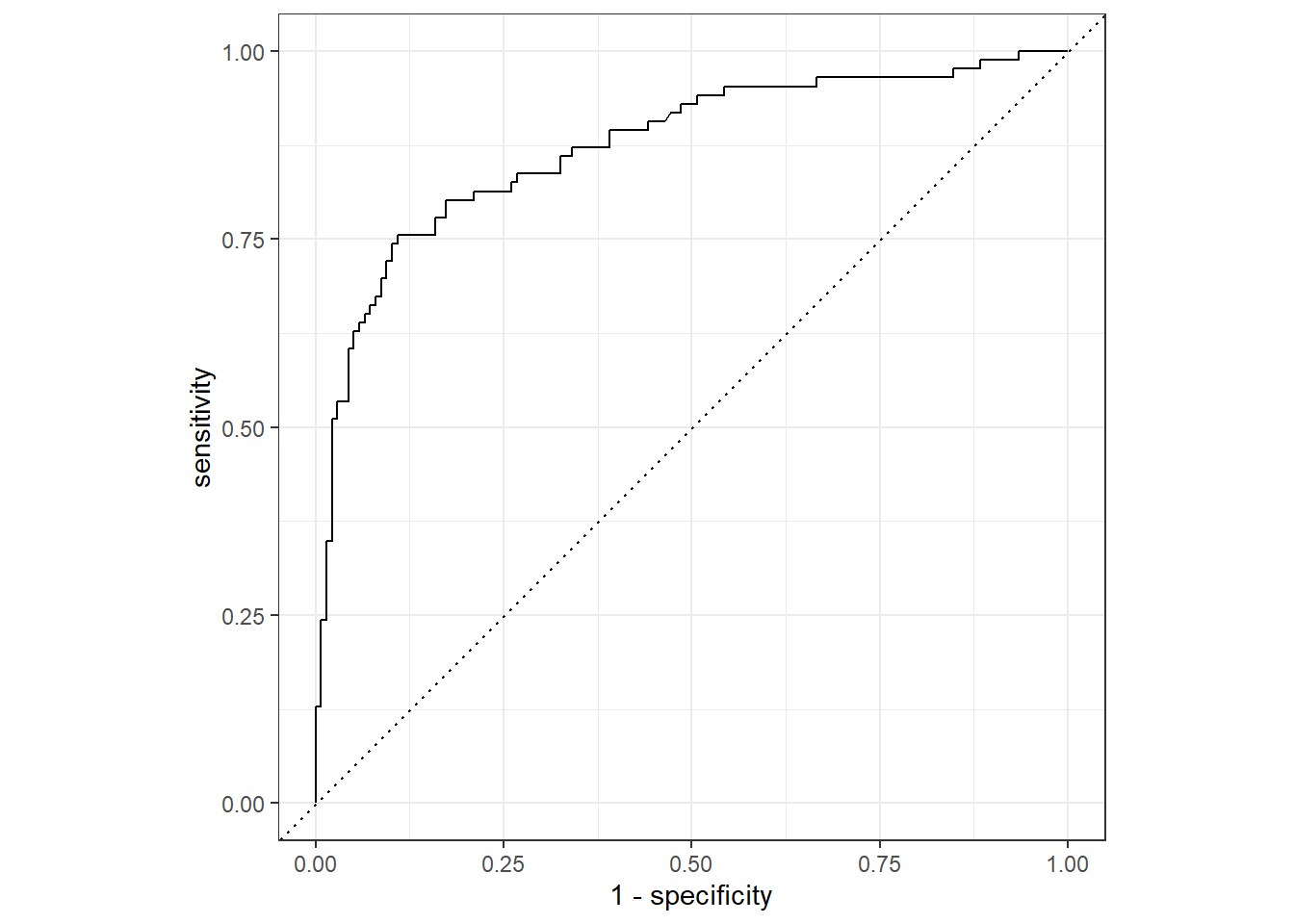
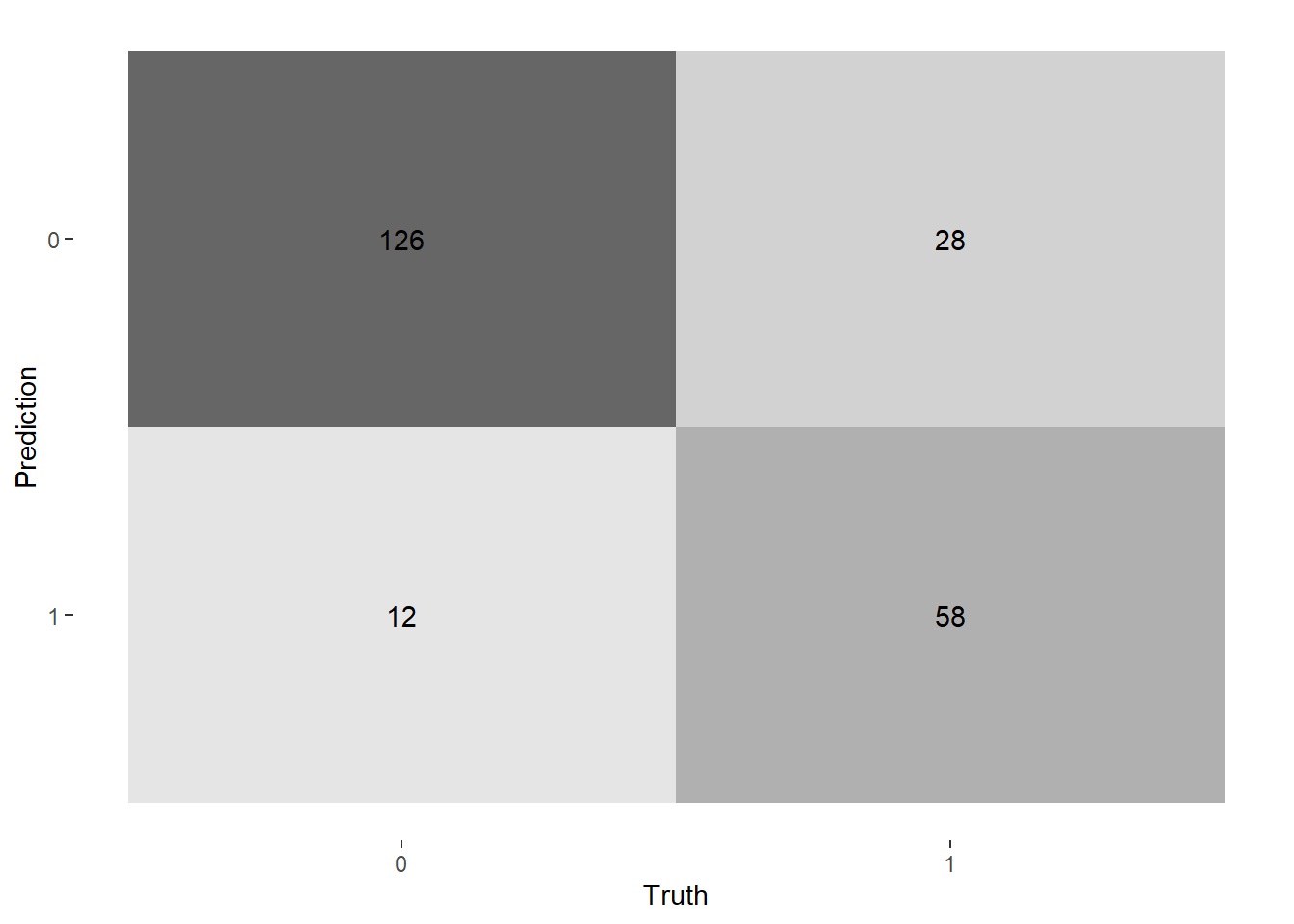
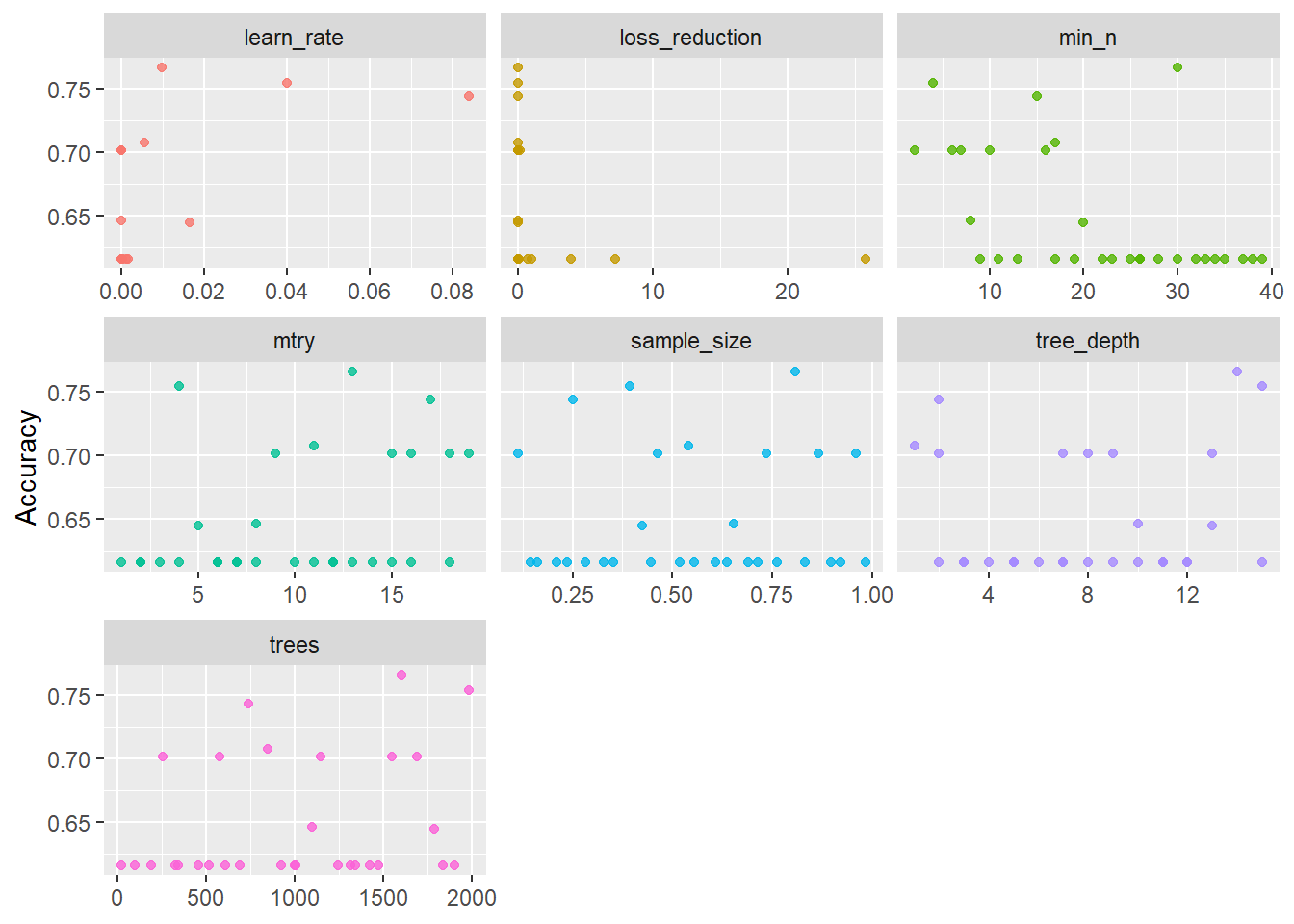
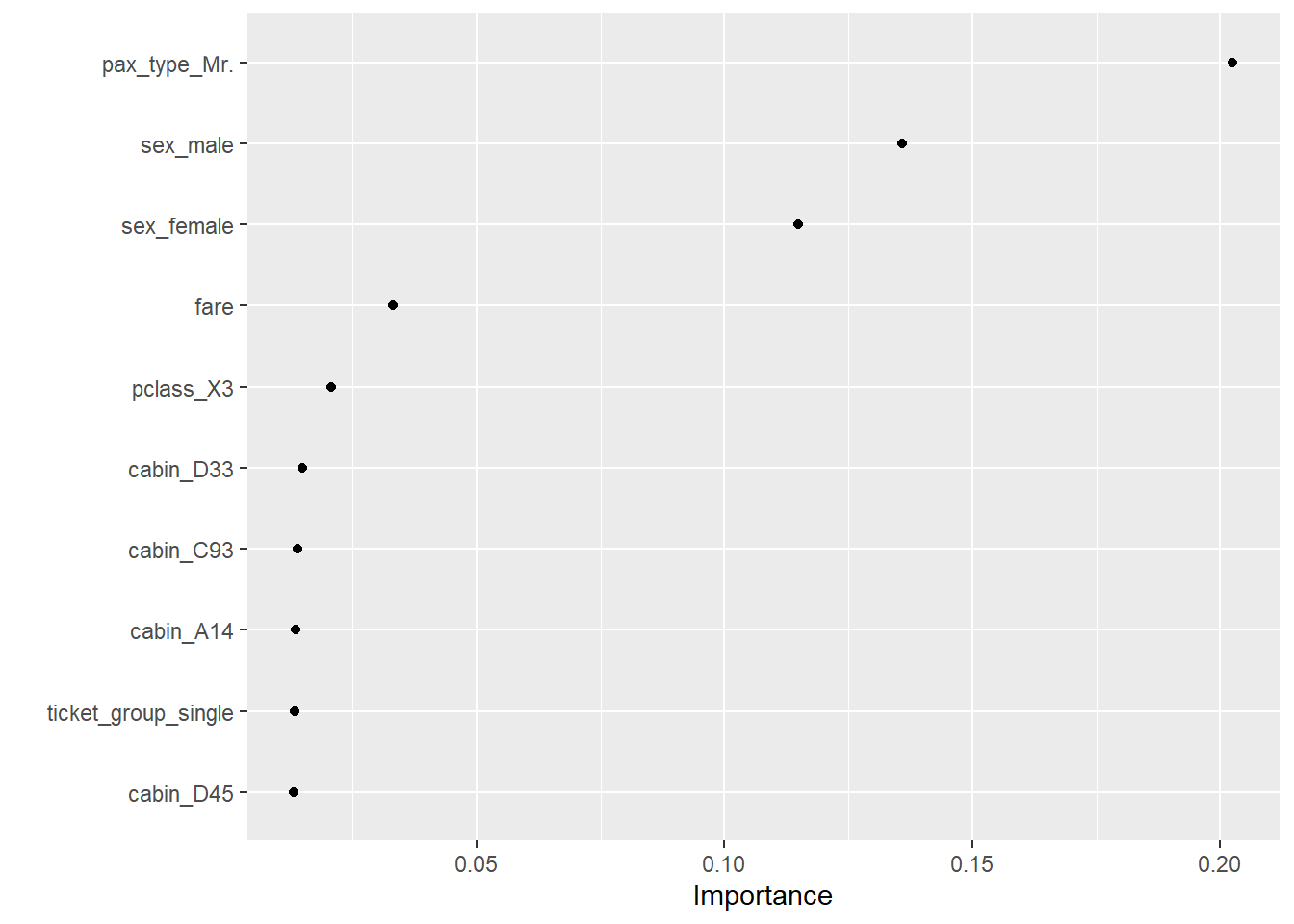
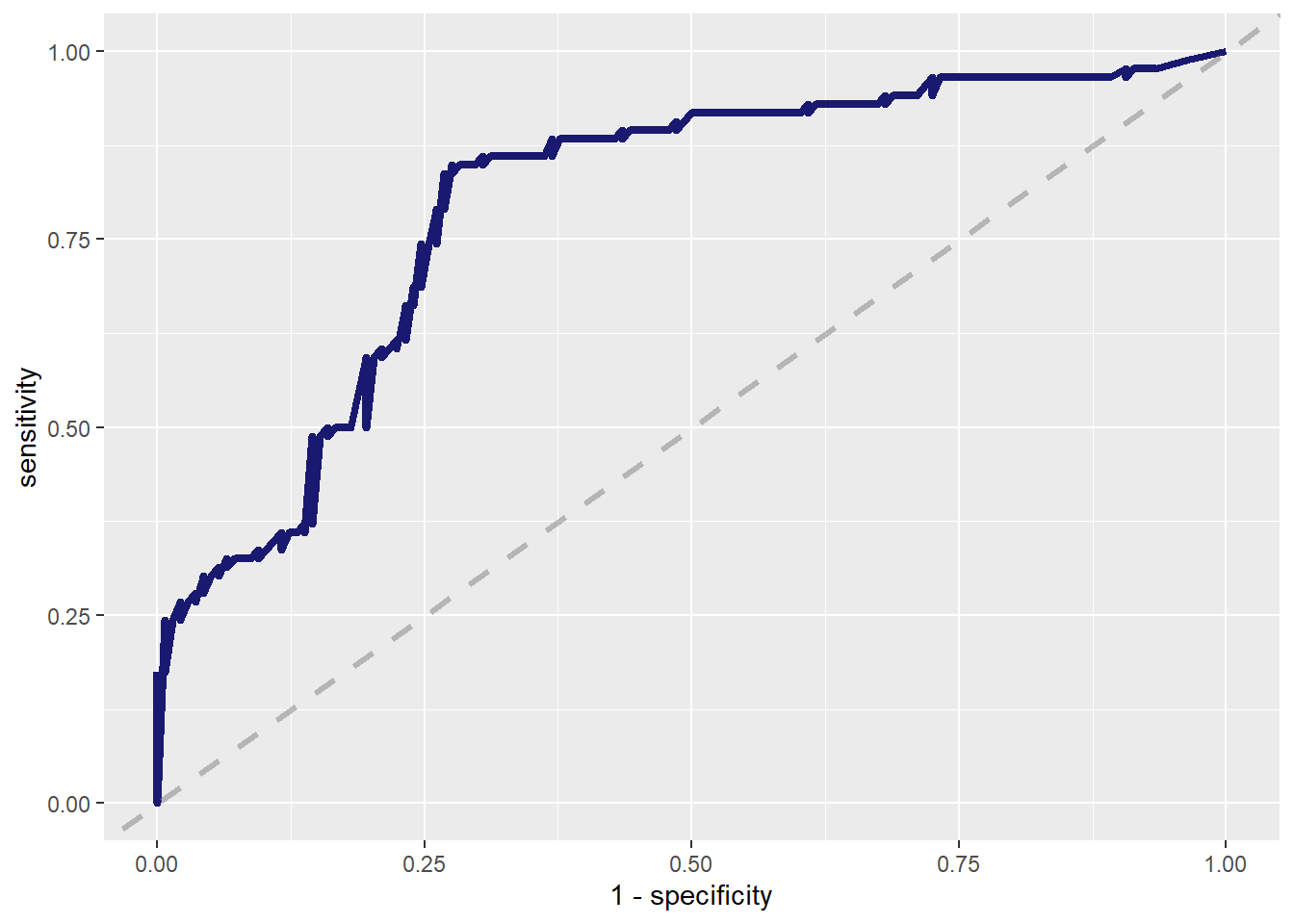
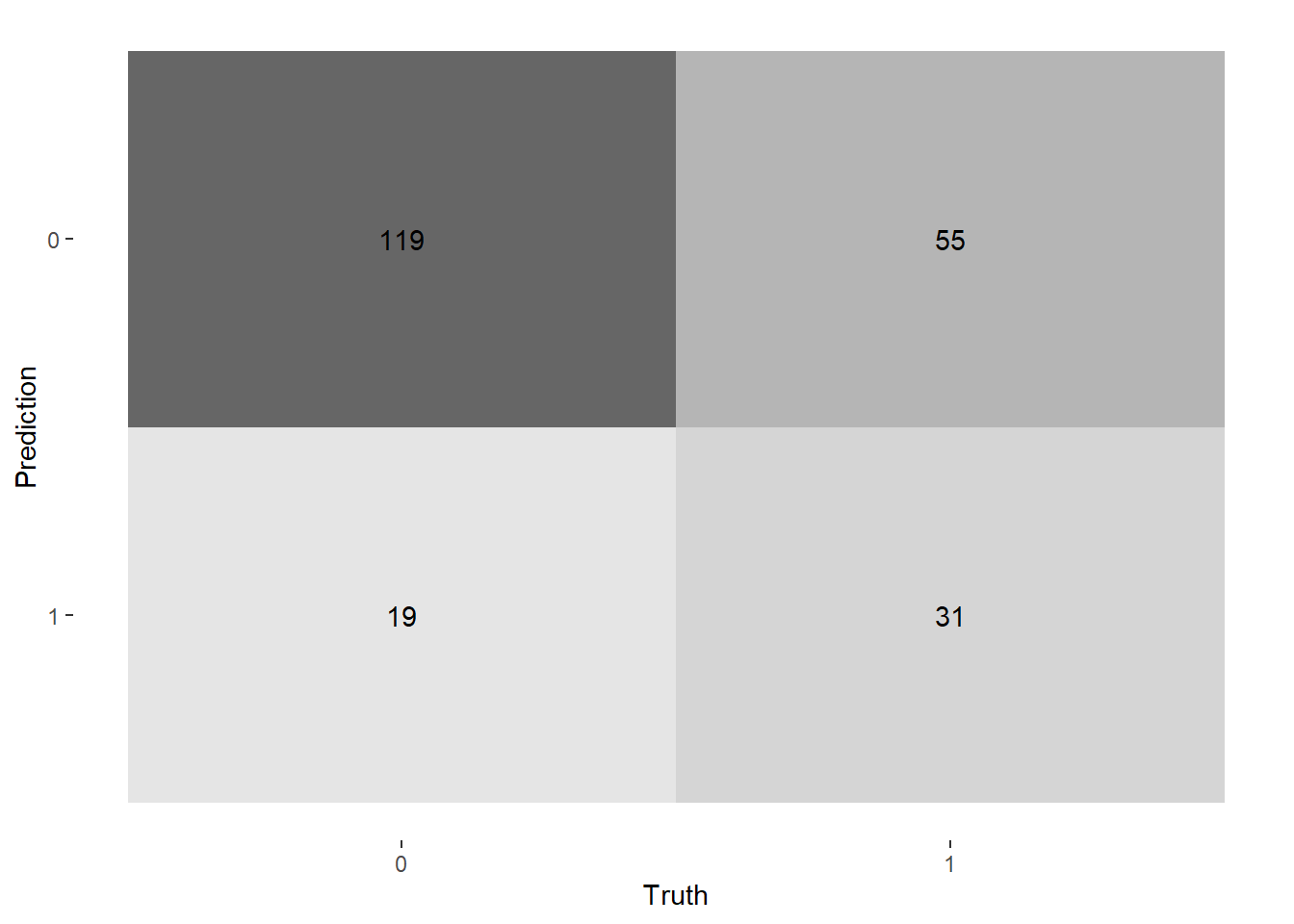
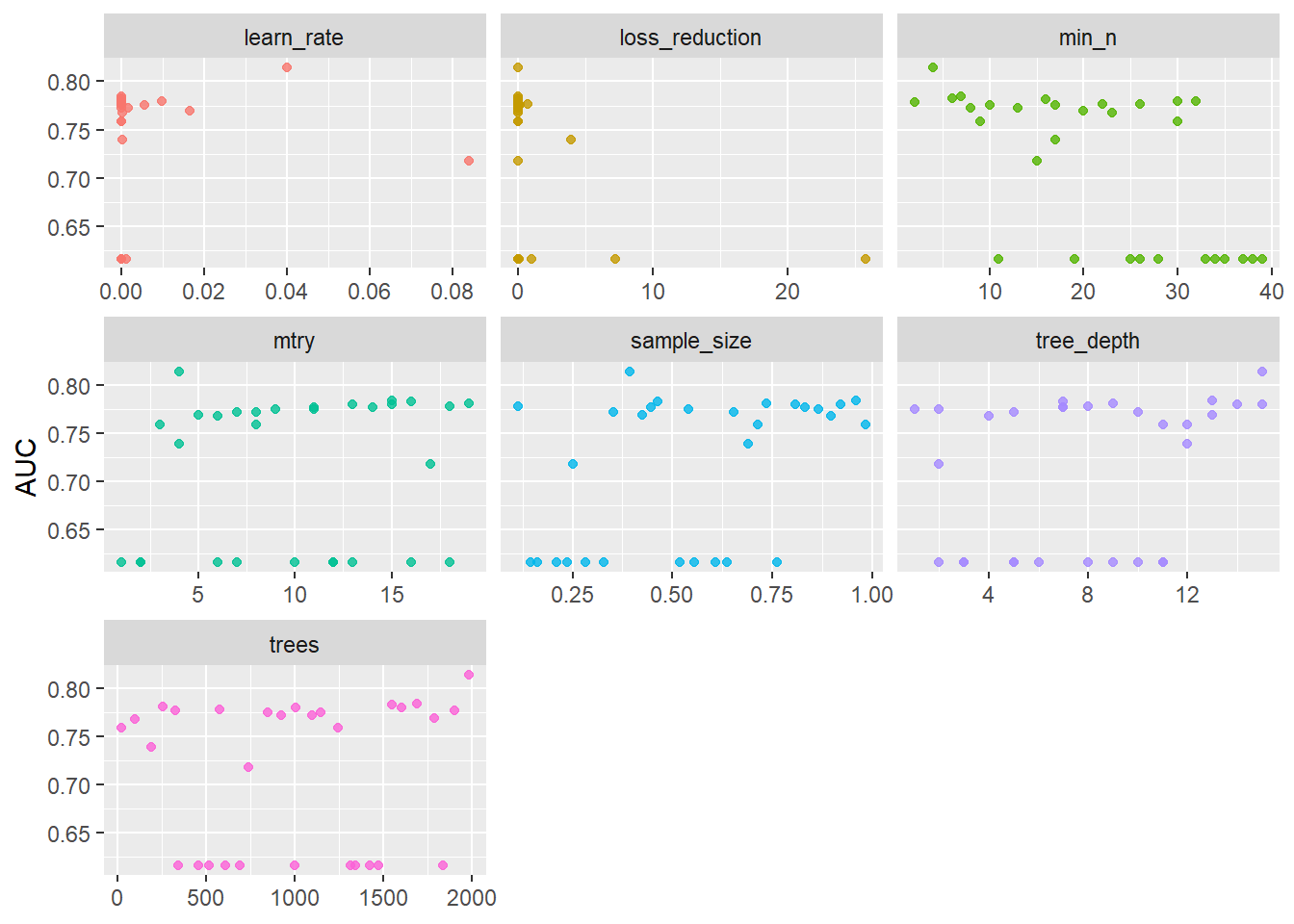
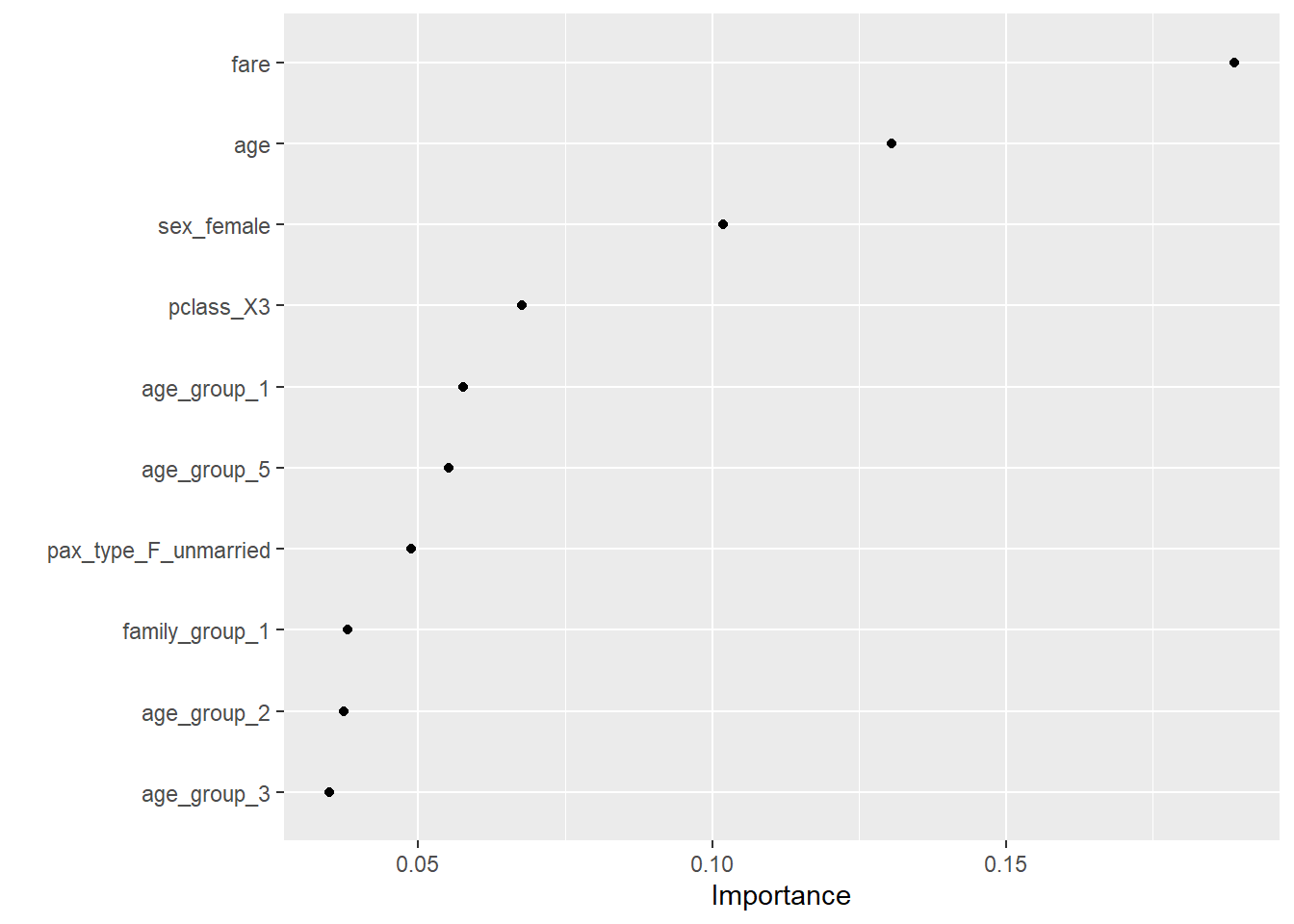
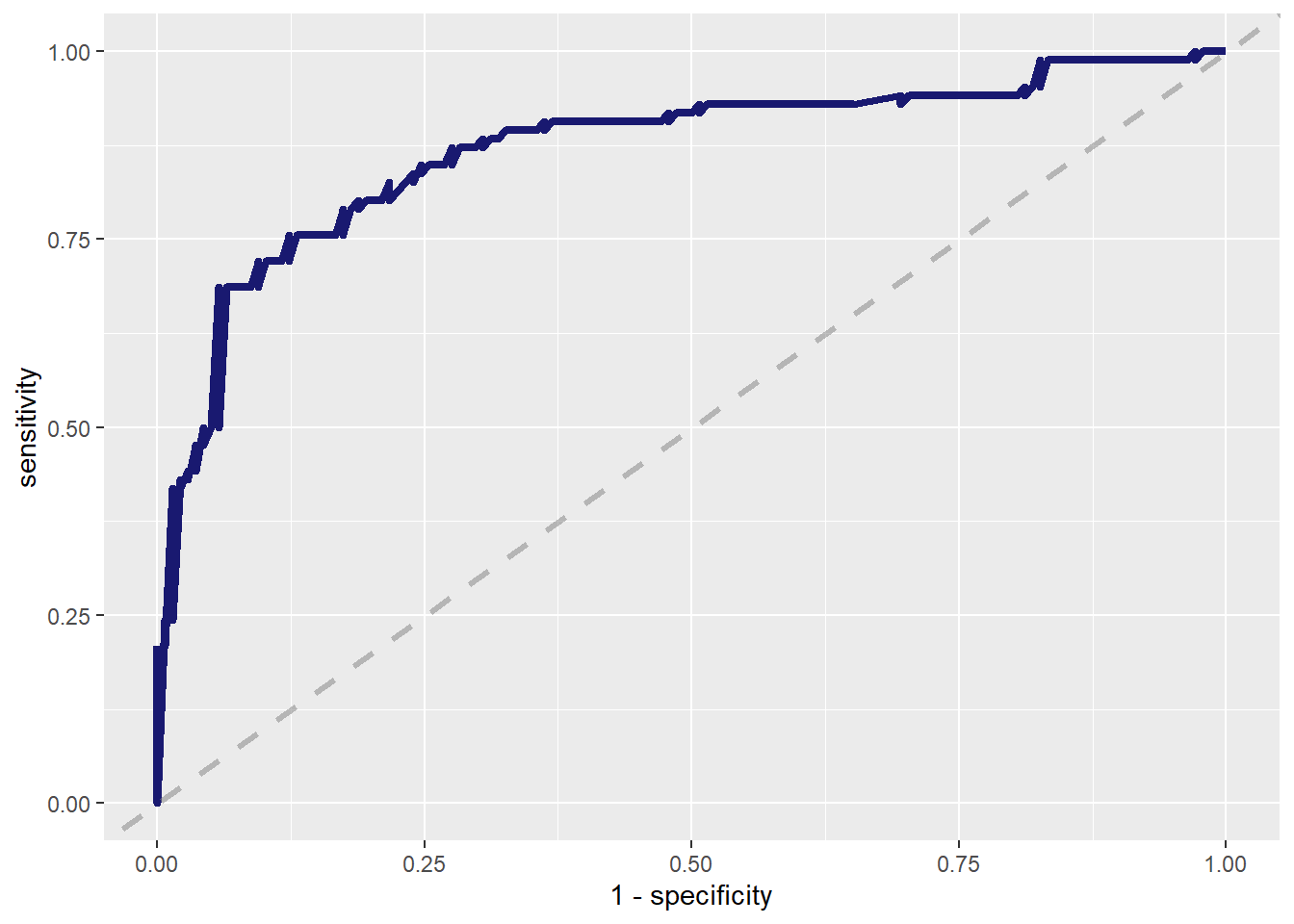
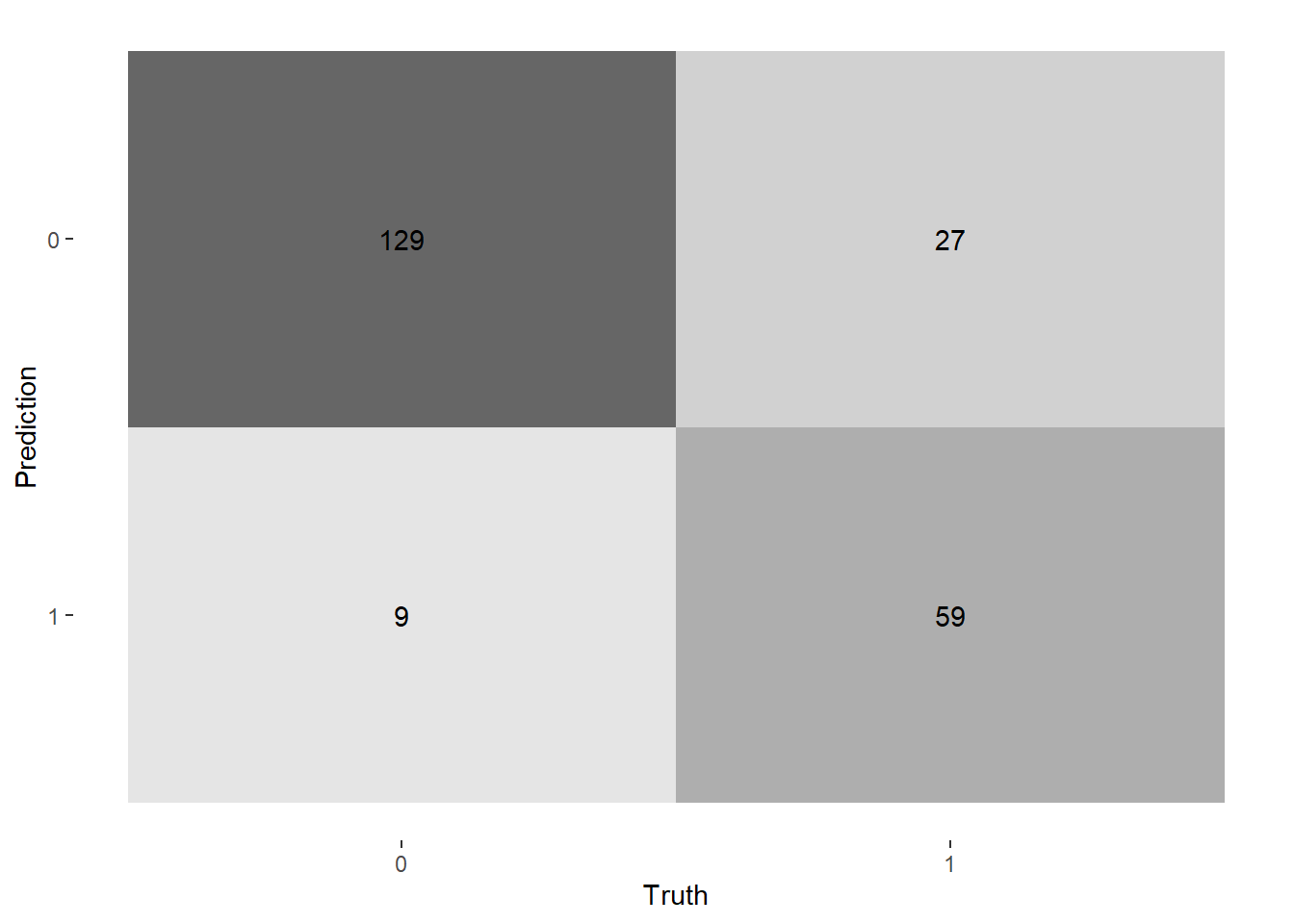
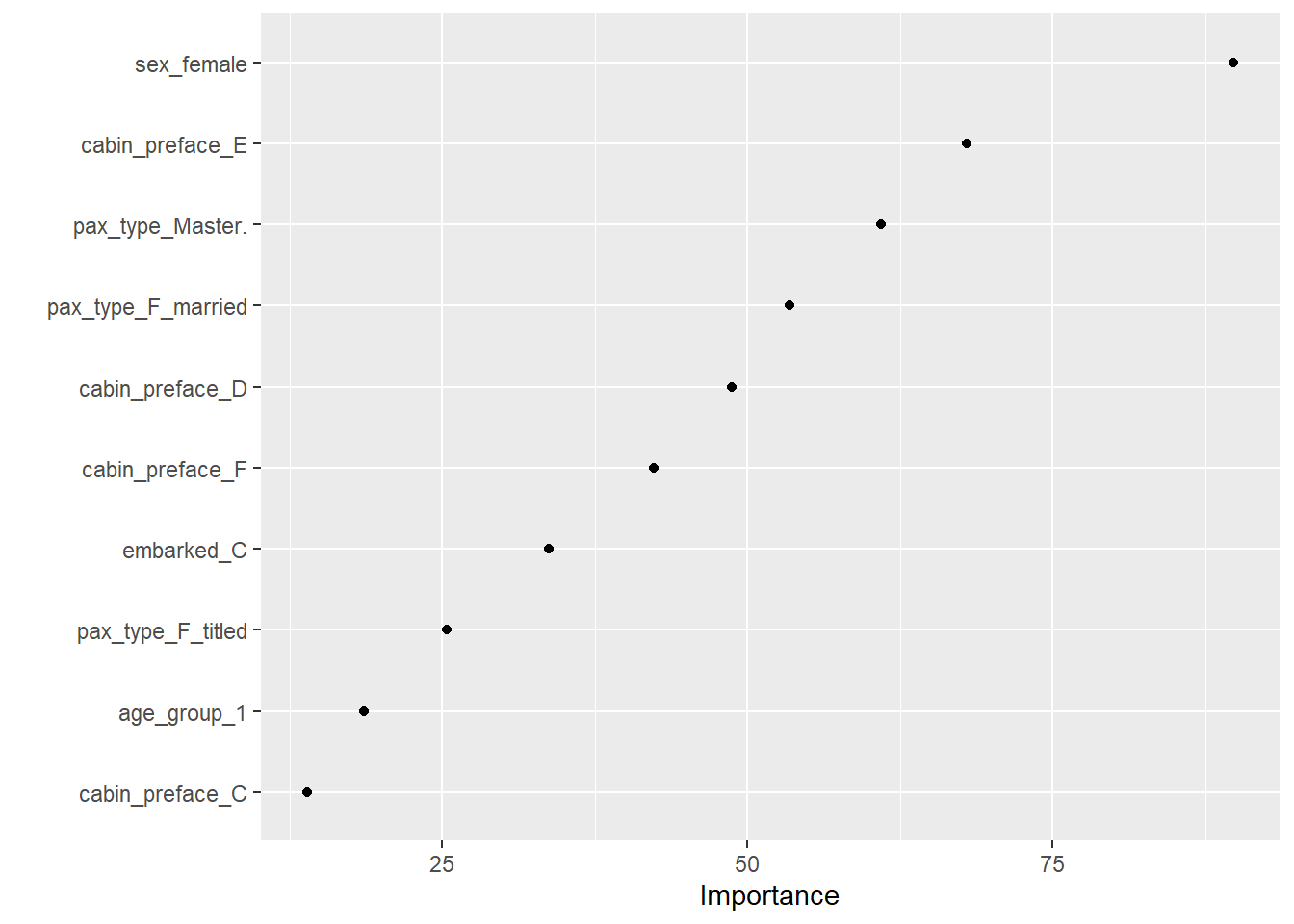
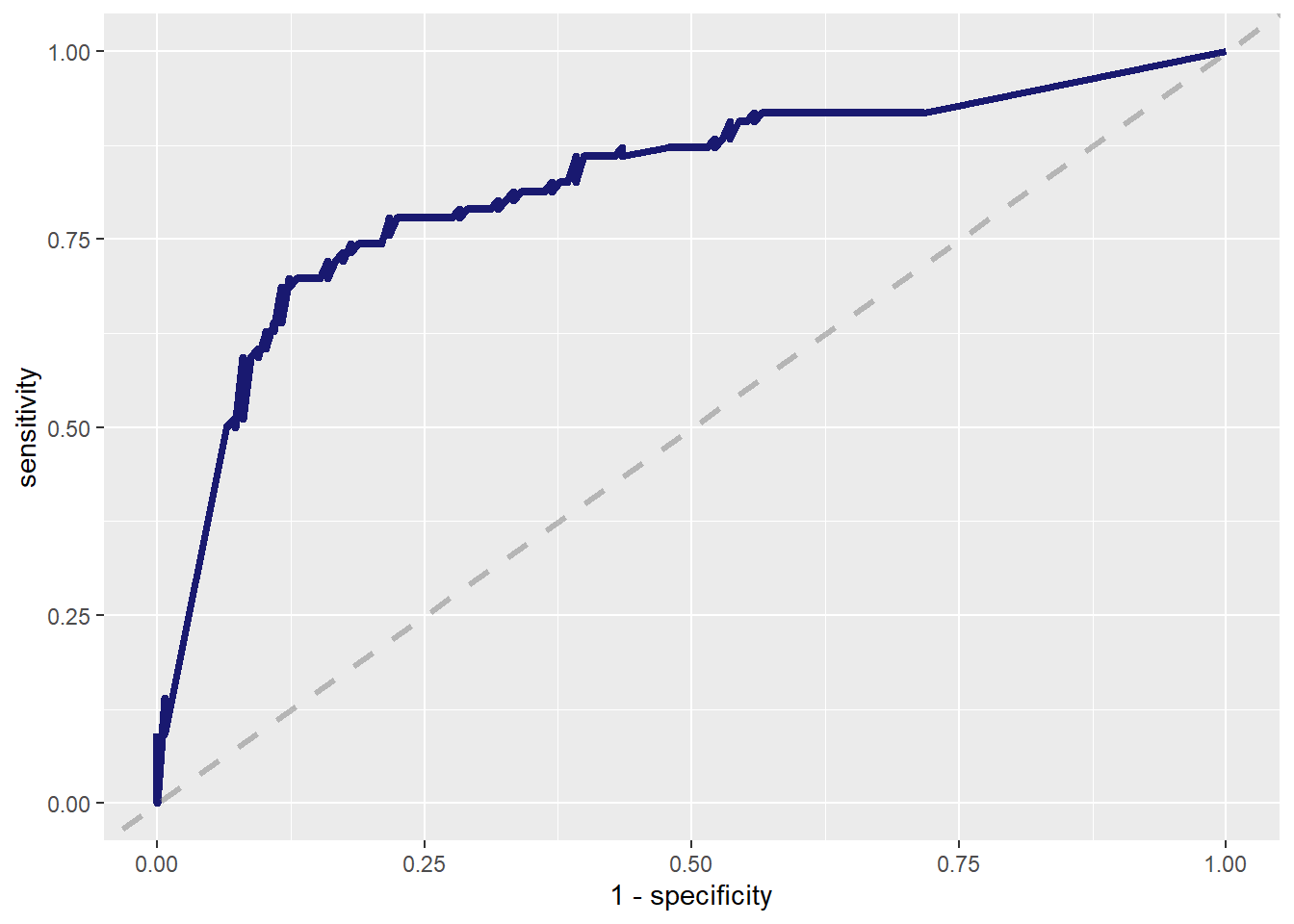
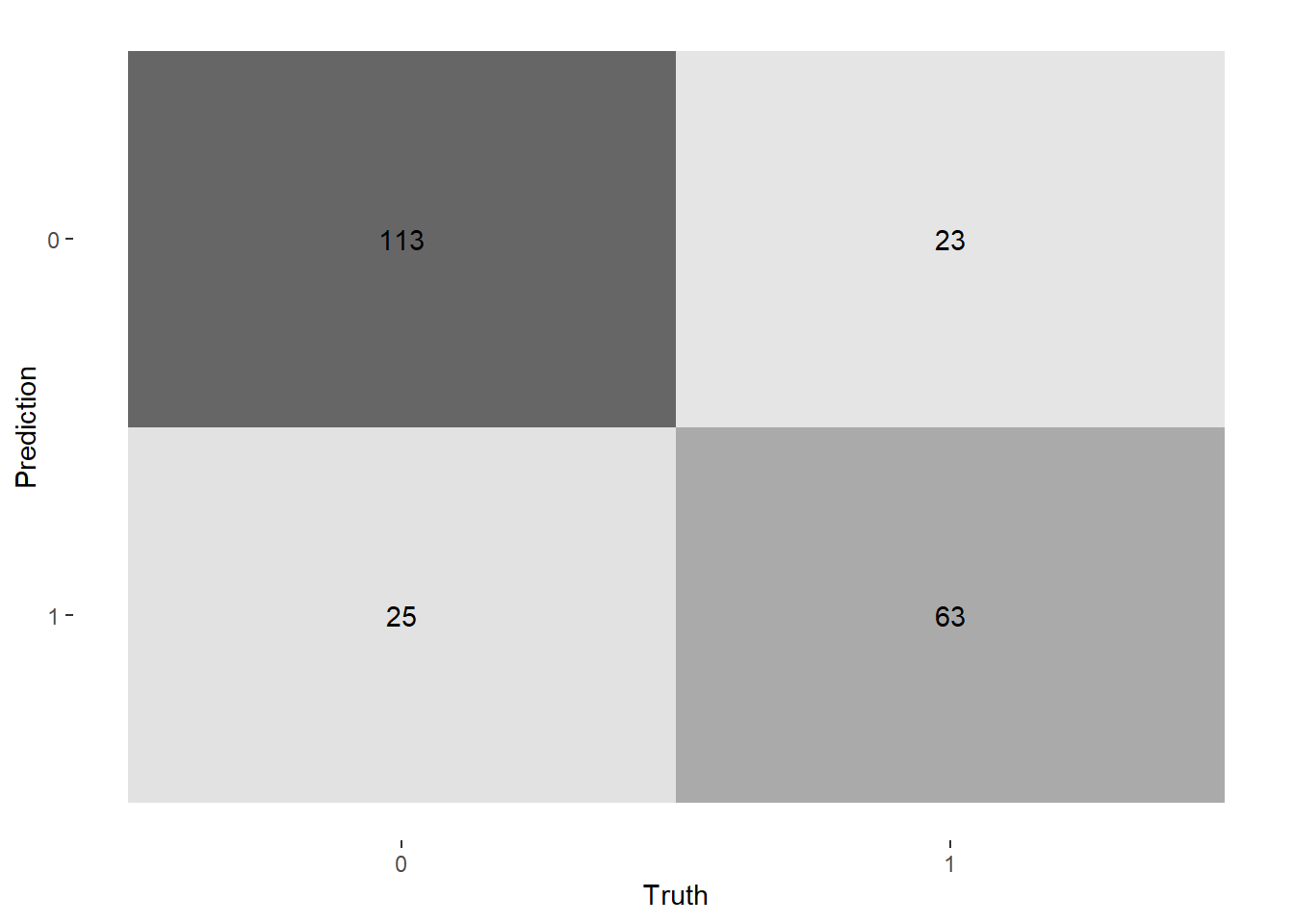
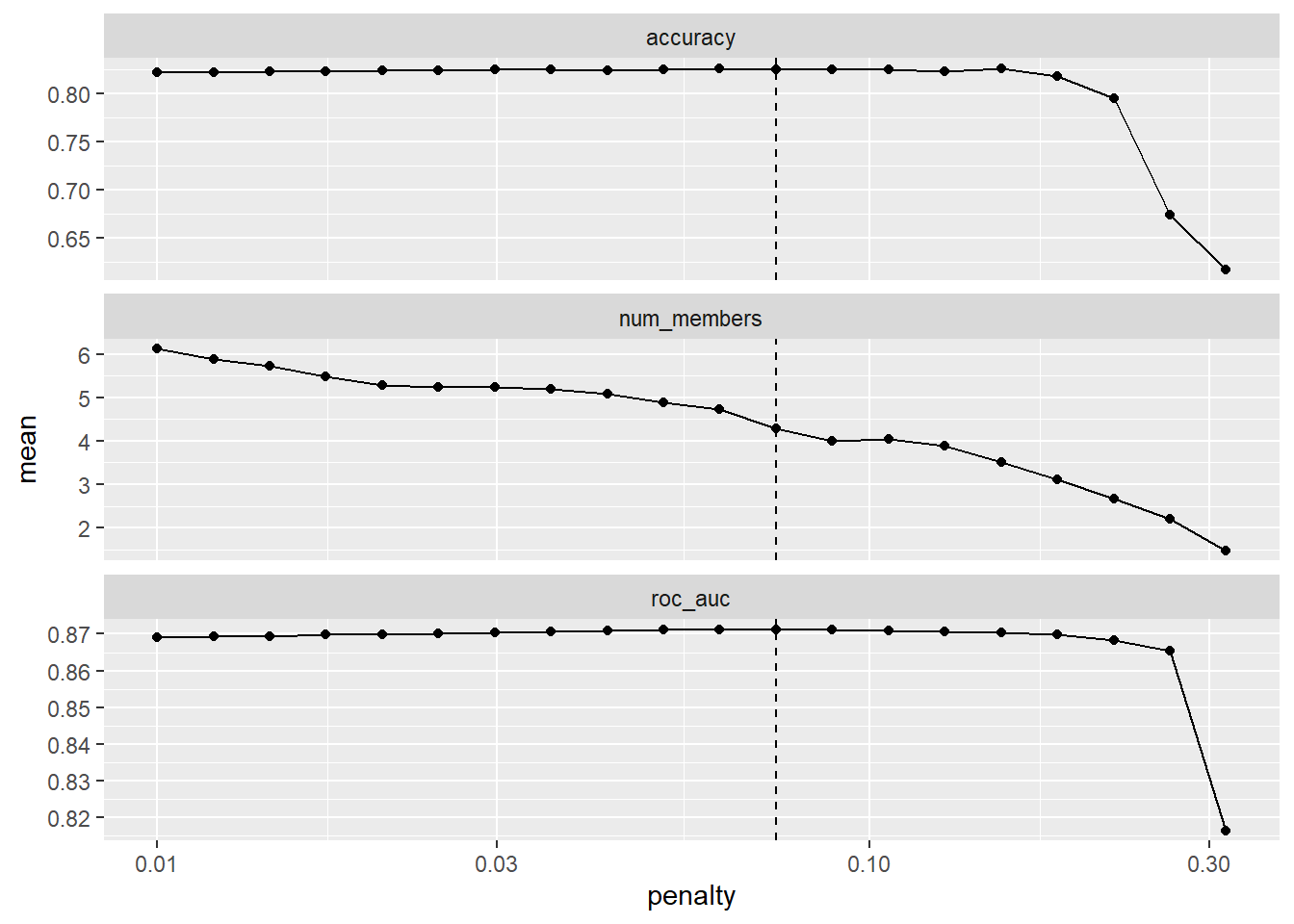
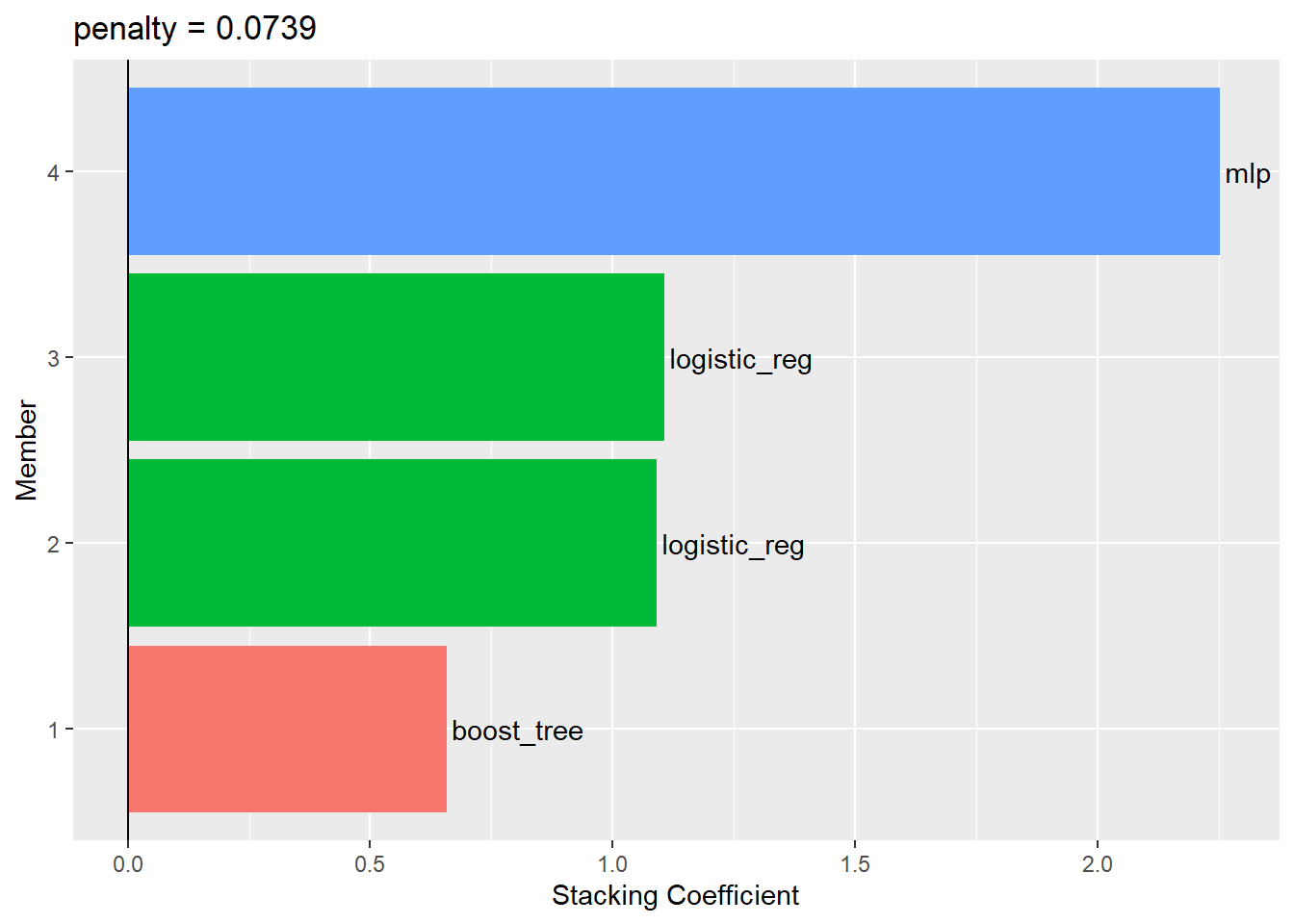
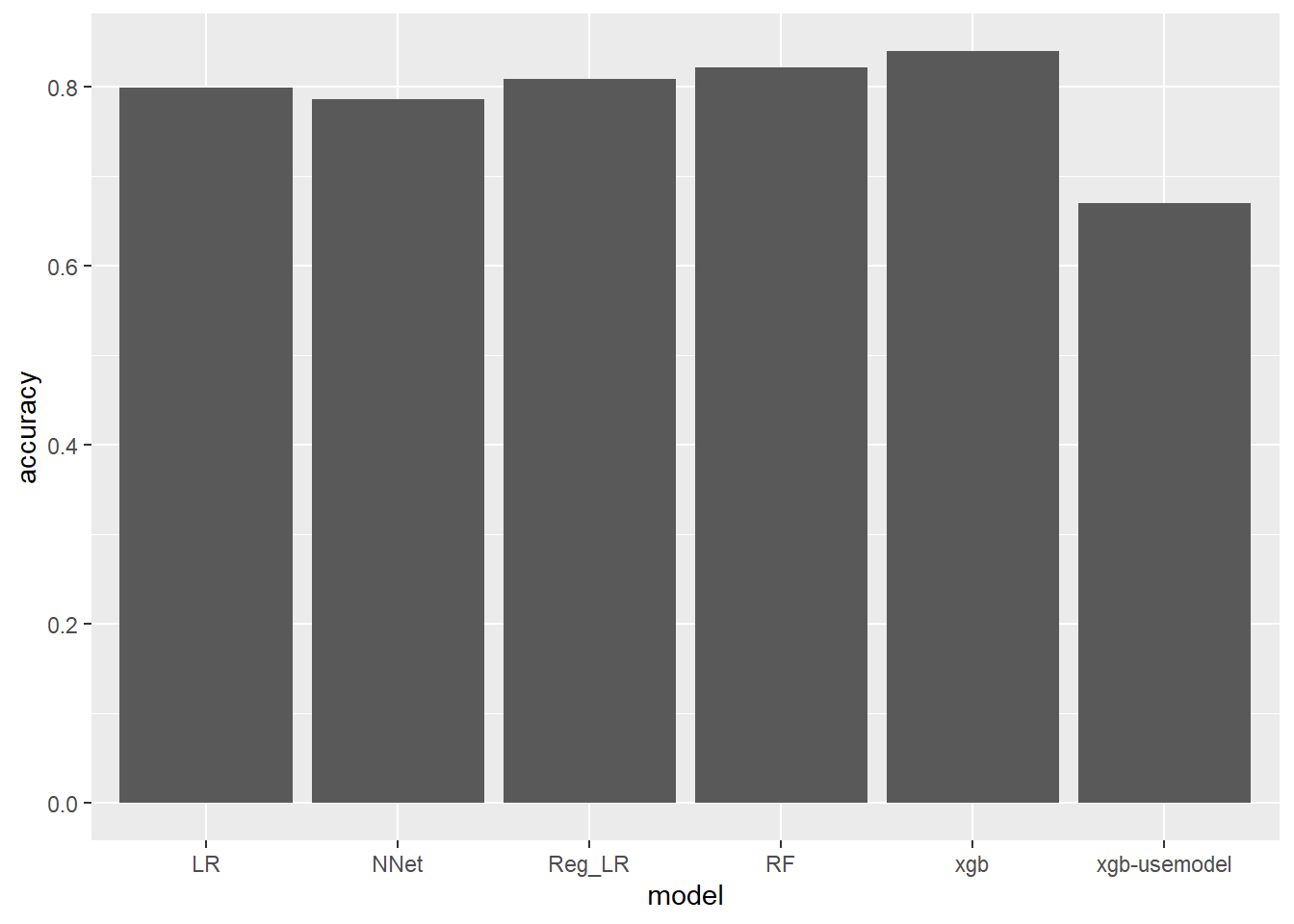
--- title: "Titanic from Kaggle" author: "Stephen Parton" date: "2022-09-01" categories: [code, analysis,titanic] website: sidebar: style: "docked" search: true contents: - section: "Data Exploration" - index.qmd format: html: theme: litera toc: true toc-title: Contents number-sections: true number-depth: 3 code-fold: true code-summary: "Code" code-tools: true execute: echo: true warning: false error: false freeze: true cache: true ---  {width="215"}## Summary ```{r, packages} #| context: setup #| include: false library (tidyverse)library (janitor)library (skimr)library (DataExplorer)library (tidymodels)library (vip)library (ggforce)tidymodels_prefer ()``` ## Final Kaggle Scores ```{r, kaggle} <- tibble (Model = c ("Logistic Regression" ,"Regularised Logistic Regression" ,"Random Forest-final" ,"Random Forest-initial" ,"XG Boost" ,"Neural Net" ,"Ensemble" ), Score = c (.76555 ,.77033 ,.77751 ,.78229 ,.77272 ,.76794 ,.77751 )%>% knitr:: kable ()``` ## Review Data ### Load Some Kaggle Data ```{r, data} #| warning: false #| echo: true #| message: false <- read_csv ("data_raw/train.csv" ,show_col_types = FALSE ) %>% clean_names () %>% mutate (train_test = "train" )<- read_csv ("data_raw/test.csv" ,show_col_types = FALSE ) %>% clean_names () %>% mutate (train_test = "test" )<- train %>% bind_rows (test)# colnames(data) # cwd() ``` ### Some Initial EDA ```{r, skim} %>% skim () ``` ### Some Initial Wrangling ```{r, wrangle_1} <- all %>% mutate (title = str_extract (name,"( \\ w)([a-z]+)( \\ .)" )) %>% mutate (pax_type = case_when (%in% c ("Miss." ,"Ms." ,"Mlle." ) ~ "F_unmarried" ,%in% c ("Mme." ,"Mrs." ) ~ "F_married" ,%in% c ("Countess." ,"Lady." ,"Dona." ) ~ "F_titled" ,%in% c ("Capt." ,"Col." ,"Major." ) ~ "Military" ,%in% c ("Dr." ,"Rev." ) ~ "M_Professional" ,%in% c ("Don." ,"Jonkheer." ,"Sir." ) ~ "M_titled" ,TRUE ~ titlesurname = str_extract (name,"( \\ w+)( \\ ,)" ),survival = ifelse (survived== 0 ,"No" ,"Yes" ),ticket_preface = str_extract (ticket,"([:graph:]+)( \\ s)" ),ticket_preface = ifelse (is.na (ticket_preface),"none" ,ticket_preface),cabin_preface = ifelse (is.na (cabin),"nk" ,substr (cabin,1 ,1 )),embarked = ifelse (is.na (embarked),"S" ,embarked)%>% group_by (pax_type,pclass) %>% mutate (age = ifelse (is.na (age),median (age,na.rm = T), age)) %>% ungroup () %>% add_count (ticket,name = "ticket_group" ) %>% mutate (ticket_group = case_when (== 1 ~ "single" ,== 2 ~ "couple" ,TRUE ~ "group" family_group = as.numeric (sib_sp)+ as.numeric (parch)+ 1 %>% mutate (family_group = factor (case_when (< 2 ~ "single" ,< 3 ~ "couple" ,TRUE ~ "family" ordered = TRUE )%>% mutate (age_group = factor (case_when (< 13 ~ "child" ,< 20 ~ "teen" ,< 30 ~ "20s" ,< 40 ~ "30s" ,< 50 ~ "40s" ,< 60 ~ "50s" ,TRUE ~ "60+" ordered = TRUE )%>% mutate (across (where (is.character),as_factor)) %>% mutate (pclass = factor (pclass,levels = c ("1" ,"2" ,"3" )),survived = factor (survived)%>% select (- c (title,ticket_preface))#all_proc %>% glimpse() ``` ### A bit more EDA ```{r, EDA_1} %>% select (- c (name,ticket,cabin,surname,train_test)) %>% :: plot_bar ()``` ```{r, data_explorer1} %>% DataExplorer:: plot_histogram (ggtheme = theme_light () )``` ### Eyeballing Survival Graphs on Training Data ```{r, eye_ball_survival, fig.height=15} #| warning: false <- all_proc %>% filter (train_test == "train" ) %>% select (passenger_id,pclass,sex,embarked,pax_type,ticket_group,family_group,age_group,cabin_preface,survival) %>% droplevels () %>% mutate (across (where (is.factor),~ factor (.x,ordered = FALSE ))) %>% pivot_longer (cols = c (pclass: cabin_preface)) <- no_f %>% split (.$ name) %>% map (~ ggplot (.,aes (y= value,fill= survival)) + geom_bar () + ggtitle (.$ name) + theme_bw () + labs (x= NULL ,y= NULL )+ scale_fill_viridis_d (option = "cividis" )library (patchwork)wrap_plots (g_l, ncol = 2 )``` ### Split Data back to Train/Test/Validation ```{r, split} <- all_proc %>% filter (train_test == "train" ) %>% select (- c (survival))<- initial_split (train_proc_adj_tbl,strata = survived)<- training (train_split)<- testing (train_split)``` ## Recipe-Base ```{r, recipe_base} <- recipe (survived ~ ., data = train_train) %>% update_role (passenger_id, name,surname,ticket,cabin,new_role = "ID" ) %>% step_impute_knn (all_numeric_predictors ()) %>% step_dummy (all_nominal_predictors ()) %>% step_factor2string (all_nominal_predictors ()) %>% step_zv (all_predictors ()) %>% step_pca ()``` ### Save Files ```{r, save_rds} write_rds (all_proc,"artifacts/all_proc.rds" )write_rds (train_split,"artifacts/train_split.rds" )write_rds (recipe_base,"artifacts/recipe_base.rds" )# # all_proc <- read_rds("artifacts/all_proc.rds") # train_split <- read_rds("artifacts/train_split.rds") # recipe_base <- read_rds("artifacts/recipe_base.rds") ``` ## Models ### Logistic Regression #### LR Model Spec ```{r, LR_model} <- logistic_reg () %>% set_engine ("glm" )``` #### LR Workflow ```{r, LR_wflow} <- workflow () %>% add_model (lr_spec) %>% add_recipe (recipe_base)``` #### LR Fit Model ```{r, LR_fit} <- %>% last_fit (train_split)#lr_fit <- lr_fit %>% collect_metrics ()#show_notes(.Last.tune.result) ``` #### LR Predict ```{r, LR_pred} <- lr_fit %>% collect_predictions () %>% rename (survived_pred = survived) %>% bind_cols (train_test)``` #### LR Performance on validation set ##### AUC Curve ```{r, LR_auc} %>% roc_curve (truth = survived,.pred_1,event_level= "second" ) %>% autoplot ()``` ##### Confusion Matrix ```{r, LR_confuse} %>% conf_mat (survived,.pred_class) %>% autoplot (type = "heatmap" )``` #### LR Resampling ```{r, LR_resample} #| message: false #| warning: false <- vfold_cv (train_train, strata = survived, v= 5 )#folds <- control_resamples (save_pred = TRUE ,save_workflow = TRUE )<- parallel:: detectCores ()<- parallel:: makePSOCKcluster (cores - 1 )# doParallel::registerDoParallel(cores = cores) set.seed (1234 )<- %>% fit_resamples (folds, control = control)#show_best(lr_fit_cv,metric= "accuracy") #lr_fit_cv <- collect_metrics (lr_fit_cv):: stopCluster (cl)``` ```{r, LR_preds} #lr_param <- extract_parameter_set_dials(lr_spec) <- collect_predictions (lr_fit_cv) %>% rename (survived_pred = survived) # bind_cols(testing(train_split)) ``` ```{r, LR_fit2} <- parallel:: makePSOCKcluster (cores - 1 )set.seed (1234 )<- lr_wflow %>% fit (data = train_proc_adj_tbl)extract_recipe (lm_fit, estimated = TRUE ):: stopCluster (cl)``` ## Regularised Logistic Regression - GLMNET ### RLR Model Spec ```{r, rlr_model} <- logistic_reg (penalty = tune (), mixture = tune ()) %>% set_engine ("glmnet" )``` ### RLR Parameter Tuning ```{r, rlr_tuning} <- extract_parameter_set_dials (rlr_model)<- grid_latin_hypercube (penalty (),mixture (),size = 30 head (rlr_grid) %>% knitr:: kable (digits = 3 )``` ### RLR Workflow ```{r, rlr_wflow} <- workflow () %>% add_model (rlr_model) %>% add_recipe (recipe_base)``` ### RLR Hyper-parameter Tuning ```{r, rlr_cvs} # rlr_folds <- vfold_cv(training(train_split), strata = survived, v=10,repeats = 5) # rlr_folds %>% tidy() #doParallel::registerDoParallel(cores = cores) <- parallel:: makePSOCKcluster (cores - 1 )set.seed (234 )<- tune_grid (resamples = folds,grid = rlr_grid,control = control_grid (save_pred = TRUE , save_workflow = TRUE )<- collect_metrics (rlr_tuning_result)head (rlr_tuning_metrics) %>% knitr:: kable (digits = 3 ):: stopCluster (cl)``` ```{r, rlr_tune} %>% collect_metrics () %>% filter (.metric == "accuracy" ) %>% select (mean, penalty,mixture) %>% pivot_longer (penalty: mixture,values_to = "value" ,names_to = "parameter" %>% ggplot (aes (value, mean, color = parameter)) + geom_point (alpha = 0.8 , show.legend = FALSE ) + facet_wrap (~ parameter, scales = "free_x" ) + labs (x = NULL , y = "AUC" )show_best (rlr_tuning_result, "accuracy" )<- select_best (rlr_tuning_result, "accuracy" )``` ### RLR Predict ```{r, rlr_predict1} <- finalize_workflow (%>% last_fit (train_split) %>% extract_fit_parsnip () %>% vip (geom = "col" )``` ```{r, rlr_predict2} <- rlr_final_wflow %>% last_fit (train_split)<- collect_metrics (rlr_final_fit)%>% knitr:: kable ()<- rlr_final_fit %>% collect_predictions ()<- rlr_test_predictions %>% bind_cols (train_test %>% select (- survived)) glimpse (rlr_test_predictions_all)# rlr_pred <- predict(rlr_final_fit,train_2 )%>% # bind_cols(predict(rlr_final_fit, train_2,type="prob")) %>% # bind_cols(train_2 %>% select(survived)) # # rlr_pred %>% # roc_auc(truth = survived, .pred_1, event_level = "second") # # rlr_pred %>% # roc_curve(truth = survived, .pred_1,event_level="second") %>% # autoplot() # # # rlr_metrics <- rlr_pred %>% # metrics(truth = survived, estimate = .pred_class) %>% # filter(.metric == "accuracy") # rlr_metrics # survive_rlr_pred <- # augment(survive_lr_fit, train_2) # survive_rlr_pred ``` ### RLR Confusion Matrix ```{r, rlr_confusion_matrix} %>% conf_mat (survived,.pred_class) %>% autoplot (type = "heatmap" )``` ## Random Forest ### RF Model Spec - Ranger ```{r, rf_model} <- rand_forest (trees = 1000 ,mtry = tune (),min_n = tune ()%>% set_engine ("ranger" ,importance = "permutation" ) %>% set_mode ("classification" )``` ### RF Workflow ```{r, rf_wflow} <- workflow () %>% add_model (rf_model) %>% add_recipe (recipe_base)``` ### RF Tuning - Initial ```{r, rf_tuning} <- parallel:: makePSOCKcluster (cores - 1 )set.seed (1234 )<- tune_grid (resamples = folds,grid = 20 :: stopCluster (cl)%>% collect_metrics () %>% filter (.metric == "accuracy" ) %>% select (mean,min_n,mtry) %>% pivot_longer (min_n: mtry) %>% ggplot (aes (value, mean, color = name)) + geom_point (show.legend = FALSE ) + facet_wrap (~ name, scales = "free_x" ) + labs (x = NULL , y = "Accuracy" )``` ```{r} <- grid_regular (mtry (range = c (5 , 40 )),min_n (range = c (5 , 30 )),levels = 5 ``` ### RF Graph Results ```{r} #| warning: false #| echo: true #| message: false #| <- parallel:: makePSOCKcluster (cores - 1 )set.seed (1234 )<- tune_grid (resamples = folds,grid = rf_grid:: stopCluster (cl)%>% collect_metrics () %>% filter (.metric == "accuracy" ) %>% mutate (min_n = factor (min_n)) %>% ggplot (aes (mtry, mean, color = min_n)) + geom_line (alpha = 0.5 , size = 1.5 ) + geom_point () + labs (y = "Accuracy" )``` ```{r} <- select_best (rf_grid_tune,"accuracy" )%>% knitr:: kable ()``` ### RF Final Model ```{r} <- finalize_model (``` ### RF Final Workflow ```{r} <- finalize_workflow (``` ### RF Parameter Importance ```{r, rf_vip} %>% fit (data = train_proc_adj_tbl) %>% extract_fit_parsnip () %>% vip (geom = "point" )``` ### RF Final Fit ```{r, rf_fit} <- %>% last_fit (train_split)<- collect_metrics (rf_final_fit)``` ### RF Predict ```{r, rf_predict} # rf_final_fit <- rf_wflow %>% fit(train_test) # class(rf_final_fit) <- collect_predictions (rf_final_fit)# fit(rf_final_wflow,train_train) %>% # predict(rf_final_wflow, new_data = train_test) %>% #bind_cols(predict(rf_final_wflow, train_test,type = "prob")) %>% #bind_cols(train_test %>% select(survived)) head (rf_test_predictions)``` ### RF Performance on Test Set ```{r, rf_perf} # rf_test_predictions %>% # roc_auc(truth = survived, .pred_1,event_level = "second") <- rf_test_predictions %>% metrics (truth = survived, estimate = .pred_class) %>% filter (.metric == "accuracy" )%>% roc_curve (truth = survived, .pred_1,event_level = "second" ) %>% autoplot ()``` ### RF Confusion Matrix ```{r, rf_confusion_matrix} %>% conf_mat (survived,.pred_class) %>% autoplot (type = "heatmap" )``` ## XG Boost - Usemodel ### XGB - Usemodel Library specs ```{r} library (usemodels)use_xgboost (survived ~ .,data= train_train,verbose = TRUE ``` ### XGB - Parameters ```{r, xgb_grid} #| warning: false #| echo: true #| message: false <- grid_latin_hypercube (tree_depth (),min_n (),trees (),loss_reduction (),sample_size = sample_prop (),finalize (mtry (), train_train),learn_rate (),size = 30 head (xgb_grid)``` ```{r, usemodel_scripts} <- recipe (formula = survived ~ ., data = train_train) %>% step_novel (all_nominal_predictors ()) %>% ## This model requires the predictors to be numeric. The most common ## method to convert qualitative predictors to numeric is to create ## binary indicator variables (aka dummy variables) from these ## predictors. However, for this model, binary indicator variables can be ## made for each of the levels of the factors (known as 'one-hot ## encoding'). step_dummy (all_nominal_predictors (), one_hot = TRUE ) %>% step_zv (all_predictors ()) <- boost_tree (trees = tune (), mtry = tune (),min_n = tune (), tree_depth = tune (), learn_rate = tune (), loss_reduction = tune (), sample_size = tune ()) %>% set_mode ("classification" ) %>% set_engine ("xgboost" ) <- workflow () %>% add_recipe (xgboost_usemodel_recipe) %>% add_model (xgboost_usemodel_model) #doParallel::registerDoParallel(cores = cores) <- parallel:: makePSOCKcluster (cores - 1 )set.seed (1234 )<- tune_grid (xgboost_usemodel_wflow, resamples = folds, grid = xgb_grid):: stopCluster (cl)``` ### XGB - Usemodel Best Parameter Settings ```{r, xgb_usemodel_para_sel} <- collect_metrics (xgboost_usemodel_tune)%>% collect_metrics () %>% filter (.metric == "accuracy" ) %>% select (mean, mtry: sample_size) %>% pivot_longer (mtry: sample_size,values_to = "value" ,names_to = "parameter" %>% ggplot (aes (value, mean, color = parameter)) + geom_point (alpha = 0.8 , show.legend = FALSE ) + facet_wrap (~ parameter, scales = "free_x" ) + labs (x = NULL , y = "Accuracy" )``` ```{r, xgb_usemodel_select_paras} show_best (xgboost_usemodel_tune, "accuracy" )<- select_best (xgboost_usemodel_tune, "accuracy" )<- finalize_workflow (``` ### XGB - Usemodel Parameter Ranking - VIP ```{r, xgb_usemodel_vip} %>% fit (data = train_train) %>% extract_fit_parsnip () %>% vip (geom = "point" )``` ### XGB - Usemodel Performance #### XGB - Usemodel Accuracy Measured on Test Set ```{r, xgb_usemodel_final_metrics} <- parallel:: makePSOCKcluster (cores - 1 )set.seed (1234 )<- last_fit (xgb_usemodel_final_wflow, train_split)<- collect_metrics (xgb_usemodel_final_res):: stopCluster (cl)``` #### XGB - Usemodel AUC on Test Set (within train) ```{r, xgb_usemodel_auc} %>% collect_predictions () %>% roc_curve ( truth = survived,.pred_1, event_level = "second" ) %>% ggplot (aes (x = 1 - specificity, y = sensitivity)) + geom_line (size = 1.5 , color = "midnightblue" ) + geom_abline (lty = 2 , alpha = 0.5 ,color = "gray50" ,size = 1.2 ``` ```{r, } <- collect_predictions (xgb_usemodel_final_res)head (xgb_usemodel_test_predictions)``` ### XGB - Usemodel Confusion Matrix ```{r} %>% conf_mat (survived,.pred_class) %>% autoplot (type = "heatmap" )``` ## XG Boost - Base Recipe ### XGB Model Spec ```{r, xgb_model} <- boost_tree (trees = tune (),tree_depth = tune (),min_n = tune (),loss_reduction = tune (),sample_size = tune (),mtry = tune (),learn_rate = tune ()) %>% set_engine ("xgboost" ) %>% set_mode ("classification" )``` ### XGB Workflow ```{r, xgb_wflow} <- workflow () %>% add_model (xgb_model) %>% add_recipe (recipe_base)``` ### XGB Hyper-Parameter Tuning ```{r} # xgb_folds <- vfold_cv(training(train_split), strata = survived) # xgb_folds #doParallel::registerDoParallel(cores = cores) set.seed (1234 )<- parallel:: makePSOCKcluster (cores - 1 )<- tune_grid (resamples = folds,grid = xgb_grid,control = control_grid (save_pred = TRUE ,save_workflow = TRUE ):: stopCluster (cl)``` ```{r} <- collect_metrics (xgb_tuning_result)%>% collect_metrics () %>% filter (.metric == "accuracy" ) %>% select (mean, mtry: sample_size) %>% pivot_longer (mtry: sample_size,values_to = "value" ,names_to = "parameter" %>% ggplot (aes (value, mean, color = parameter)) + geom_point (alpha = 0.8 , show.legend = FALSE ) + facet_wrap (~ parameter, scales = "free_x" ) + labs (x = NULL , y = "AUC" )``` #### XGB Best Parameters then Finalise Workflow ```{r} show_best (xgb_tuning_result, "accuracy" )<- select_best (xgb_tuning_result, "accuracy" )<- finalize_workflow (``` ```{r} %>% fit (data = train_train) %>% extract_fit_parsnip () %>% vip (geom = "point" )``` ### XGB Performance on Training Test Set #### XGB Accuracy Measured on Test Set ```{r} <- last_fit (xgb_final_wflow, train_split)<- collect_metrics (xgb_final_res)``` #### XGB AUC on Test Set (within train) ```{r} %>% collect_predictions () %>% roc_curve ( truth = survived,.pred_1, event_level = "second" ) %>% ggplot (aes (x = 1 - specificity, y = sensitivity)) + geom_line (size = 1.5 , color = "midnightblue" ) + geom_abline (lty = 2 , alpha = 0.5 ,color = "gray50" ,size = 1.2 ``` ```{r} <- collect_predictions (xgb_final_res)head (xgb_test_predictions)``` ### XGB Confusion Matrix ```{r} %>% conf_mat (survived,.pred_class) %>% autoplot (type = "heatmap" )``` ## Neural Net ### NN Model ```{r, nn_model} <- mlp (hidden_units = tune (), penalty = tune (), epochs = tune ()) %>% set_engine ("nnet" , MaxNWts = 2600 ) %>% set_mode ("classification" )%>% translate ()``` ### NN Workflow ```{r, nn_wflow} <- workflow () %>% add_model (nnet_model) %>% add_recipe (recipe_base)``` ### NN Parameters ```{r, nn_params} <- grid_latin_hypercube (hidden_units (),penalty (),epochs ()head (nnet_grid) ``` ### NN Hyper-Parameter Tuning ```{r, nn_tuning} # nnet_folds <- vfold_cv(train_train, strata = survived) # nnet_folds # doParallel::registerDoParallel(cores = cores) <- parallel:: makePSOCKcluster (cores - 1 )set.seed (1234 )<- tune_grid (resamples = folds,grid = nnet_grid,control = control_grid (save_pred = TRUE ,save_workflow = TRUE ):: stopCluster (cl)``` ### NN Best Parameters and Finalise Workflow ```{r, nn_best_params} show_best (nnet_tuning_result, "accuracy" )<- select_best (nnet_tuning_result, "accuracy" )<- select_best (xgb_tuning_result, "accuracy" )<- finalize_workflow (``` ```{r, nn_final_train} %>% fit (data = train_train) %>% extract_fit_parsnip () %>% vip (geom = "point" )``` ### NN Accuracy - Train/Test Set ```{r} <- collect_metrics (nnet_tuning_result)<- last_fit (nnet_final_wflow, train_split)<- collect_metrics (nnet_final_res)``` ### NN AUC ```{r} %>% collect_predictions () %>% roc_curve ( truth = survived,.pred_1, event_level = "second" ) %>% ggplot (aes (x = 1 - specificity, y = sensitivity)) + geom_line (size = 1.5 , color = "midnightblue" ) + geom_abline (lty = 2 , alpha = 0.5 ,color = "gray50" ,size = 1.2 ``` ### NN Predictions on Train/Test Set ```{r} <- nnet_final_res %>% collect_predictions () head (nnet_test_predictions)``` ### NN Confusion Matrix ```{r, NN_confusion_matrix} %>% conf_mat (survived,.pred_class) %>% autoplot (type = "heatmap" )``` ## Stack Models ### Stack Recipe ```{r, stack_recipe} <- recipe (survived ~ ., data = train_train) %>% update_role (passenger_id, name,surname,ticket,cabin,new_role = "ID" ) %>% step_impute_knn (all_numeric_predictors ()) %>% step_dummy (all_nominal_predictors ()) %>% step_factor2string (all_nominal_predictors ()) %>% step_zv (all_predictors ()) %>% step_pca ()<- prep (recipe_base)``` ### Stack Controls ```{r, stack_controls} <- control_resamples (save_pred = TRUE , save_workflow = TRUE )#stack_folds <- vfold_cv(training(train_split), v=10,strata = "survived") library (stacks)<- stacks () %>% #add_candidates(lr_wflow) %>% #add_candidates(rf_wflow) %>% add_candidates (nnet_tuning_result) %>% add_candidates (rlr_tuning_result) %>% add_candidates (xgb_tuning_result)``` ### Stack Blend ```{r} <- parallel:: makePSOCKcluster (cores - 1 )set.seed (1234 )<- blend_predictions (model_stack,penalty = 10 ^ seq (- 2 , - 0.5 , length = 20 ))autoplot (ensemble):: stopCluster (cl)``` ```{r,ensemble_table} ``` ### Stack Weights ```{r} autoplot (ensemble, "weights" ) + geom_text (aes (x = weight + 0.01 , label = model), hjust = 0 ) + theme (legend.position = "none" ) ``` ### Fit Member Models ```{r, fit_ensemble} <- fit_members (ensemble)collect_parameters (ensemble,"xgb_tuning_result" )``` ### Stack Predict ```{r} #ensemble_metrics <- metric_set(roc_auc,accuracy) <- predict (ensemble,train_test) %>% bind_cols (train_test) # ensemble_test_predictions <- ensemble_test_predictions %>% # mutate(.pred_class=as.numeric(.pred_class)) %>% # mutate(survived =as.numeric(survived)) # # ensemble_test_predictions <- ensemble_test_predictions %>% # mutate(roc = roc_auc(truth=survived, estimate = .pred_class)) glimpse (ensemble_test_predictions)``` ## Join Model Prediction Data ```{r, all_predictions} <- %>% mutate (model = "LR" ) %>% bind_rows (nnet_test_predictions %>% mutate (model = "NNet" )) %>% bind_rows (rlr_test_predictions %>% mutate (model = "Reg_LR" )) %>% bind_rows (rf_test_predictions %>% mutate (model = "RF" )) %>% bind_rows (xgb_test_predictions %>% mutate (model = "xgb" )) %>% bind_rows (xgb_usemodel_test_predictions %>% mutate (model = "xgb_usemodel" )) %>% bind_rows (ensemble_test_predictions %>% mutate (model = "ensemble" ))%>% head () %>% knitr:: kable ()``` ## All Metrics ```{r, all_metrics} <- %>% mutate (model = "LR" ) %>% bind_rows (nnet_final_metrics %>% mutate (model = "NNet" )) %>% bind_rows (rlr_final_metrics %>% mutate (model = "Reg_LR" )) %>% bind_rows (rf_final_metrics %>% mutate (model = "RF" )) %>% bind_rows (xgb_final_metrics %>% mutate (model = "xgb" )) %>% bind_rows (xgb_usemodel_final_metrics %>% mutate (model = "xgb-usemodel" )) <- all_metrics %>% pivot_wider (names_from = .metric,values_from = .estimate) %>% arrange (desc (accuracy))write_rds (all_metrics,"artifacts/all_metrics.rds" )%>% knitr:: kable (digits= 3 )``` ```{r, graph_all_metrics} %>% filter (.metric == "accuracy" ) %>% select (model, accuracy = .estimate) %>% ggplot (aes (model, accuracy)) + geom_col ()``` # Final Submission ```{r, predict_test} # all_predictions %>% # distinct(model) <- all_proc %>% filter (train_test== "test" )# LR ---- <- %>% fit (train_proc_adj_tbl) %>% predict (new_data= test_proc) %>% bind_cols (test_proc)<- final_test_pred_LR %>% select (PassengerID = passenger_id,Survived = .pred_class)write_csv (submission_LR,"titanic_submission_LR.csv" ) # RLR ---- <- %>% fit (train_proc_adj_tbl) %>% predict (new_data= test_proc) %>% bind_cols (test_proc)<- final_test_pred_RLR %>% select (PassengerID = passenger_id,Survived = .pred_class)write_csv (submission_RLR,"titanic_submission_RLR.csv" ) # RF ---- <- %>% fit (train_proc_adj_tbl) %>% predict (new_data= test_proc) %>% bind_cols (test_proc)<- final_test_pred_RF %>% select (PassengerID = passenger_id,Survived = .pred_class)write_csv (submission_RF,"titanic_submission_RF.csv" ) # NN ---- <- %>% fit (train_proc_adj_tbl) %>% predict (new_data= test_proc) %>% bind_cols (test_proc)<- final_test_pred_NN %>% select (PassengerID = passenger_id,Survived = .pred_class)write_csv (submission_NN,"titanic_submission_NN.csv" ) # XGB ----- <- %>% fit (train_proc_adj_tbl) %>% predict (new_data= test_proc) %>% bind_cols (test_proc)<- final_test_pred_xgb %>% select (PassengerID = passenger_id,Survived = .pred_class)write_csv (submission_xgb,"titanic_submission_xgb.csv" )# ensemble ----- <- %>% #fit(train_proc_adj_tbl) %>% predict (new_data= test_proc) %>% bind_cols (test_proc)<- final_test_pred_ens %>% select (PassengerID = passenger_id,Survived = .pred_class)write_csv (submission_ens,"titanic_submission_ens.csv" )```